ADAR enzyme and miRNA story: a nucleotide that can make the difference
- PMID: 24256817
- PMCID: PMC3856091
- DOI: 10.3390/ijms141122796
ADAR enzyme and miRNA story: a nucleotide that can make the difference
Abstract
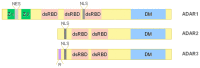
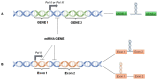
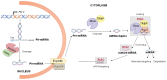
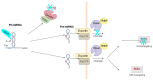
Adenosine deaminase acting on RNA (ADAR) enzymes convert adenosine (A) to inosine (I) in double-stranded (ds) RNAs. Since Inosine is read as Guanosine, the biological consequence of ADAR enzyme activity is an A/G conversion within RNA molecules. A-to-I editing events can occur on both coding and non-coding RNAs, including microRNAs (miRNAs), which are small regulatory RNAs of ~20-23 nucleotides that regulate several cell processes by annealing to target mRNAs and inhibiting their translation. Both miRNA precursors and mature miRNAs undergo A-to-I RNA editing, affecting the miRNA maturation process and activity. ADARs can also edit 3' UTR of mRNAs, further increasing the interplay between mRNA targets and miRNAs. In this review, we provide a general overview of the ADAR enzymes and their mechanisms of action as well as miRNA processing and function. We then review the more recent findings about the impact of ADAR-mediated activity on the miRNA pathway in terms of biogenesis, target recognition, and gene expression regulation.
Figures
References
-
- Lee J.T. Epigenetic regulation by long noncoding RNAs. Science. 2012;338:1435–1439. - PubMed
-
- Baltimore D. Our genome unveiled. Nature. 2001;409:814–816. - PubMed
-
- Gott J.M., Emeson R.B. Functions and mechanisms of RNA editing. Annu. Rev. Genet. 2000;34:499–531. - PubMed
-
- Gallo A., Locatelli F. ADARs: Allies or enemies? The importance of A-to-I RNA editing in human disease: From cancer to HIV-1. Biol. Rev. Camb. Philos. Soc. 2011;87:95–110. - PubMed
-
- Keegan L.P., Gallo A., O’Connell M.A. The many roles of an RNA editor. Nat. Rev. Genet. 2001;2:869–878. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

