Improved workflows for high throughput library preparation using the transposome-based Nextera system
- PMID: 24256843
- PMCID: PMC4222894
- DOI: 10.1186/1472-6750-13-104
Improved workflows for high throughput library preparation using the transposome-based Nextera system
Abstract
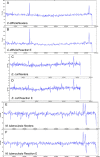
Background: The Nextera protocol, which utilises a transposome based approach to create libraries for Illumina sequencing, requires pure DNA template, an accurate assessment of input concentration and a column clean-up that limits its applicability for high-throughput sample preparation. We addressed the identified limitations to develop a robust workflow that supports both rapid and high-throughput projects also reducing reagent costs.
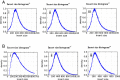
Results: We show that an initial bead-based normalisation step can remove the need for quantification and improves sample purity. A 75% cost reduction was achieved with a low-volume modified protocol which was tested over genomes with different GC content to demonstrate its robustness. Finally we developed a custom set of index tags and primers which increase the number of samples that can simultaneously be sequenced on a single lane of an Illumina instrument.
Conclusions: We addressed the bottlenecks of Nextera library construction to produce a modified protocol which harnesses the full power of the Nextera kit and allows the reproducible construction of libraries on a high-throughput scale reducing the associated cost of the kit.
Figures


References
-
- Adey A, Morrison H, Asan G, Xun X, Kitzman J, Turner E, Stackhouse B, MacKenzie A, Caruccio N, Zhang X, Shendure J. Rapid, low-input, low-bias construction of shotgun fragment libraries by high-density in vitro transposition. Genome Biol. 2010;11:R119. doi: 10.1186/gb-2010-11-12-r119. - DOI - PMC - PubMed
-
- Caruccio N. Preparation of next-generation sequencing libraries using Nextera technology: simultaneous DNA fragmentation and adaptor tagging by in vitro transposition. Methods Mol Biol. 2011;733:241–255. - PubMed
-
- Marine R, Polson SW, Ravel J, Hatfull G, Russell D, Sullivan M, Syed F, Dumas M, Wommack KE. Evaluation of a transposase protocol for rapid generation of shotgun high-throughput sequencing libraries from nanogram quantities of DNA. Appl Environ Microbiol. 2011;77:8071–8079. doi: 10.1128/AEM.05610-11. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

