The S40 residue in HIV-1 Gag p6 impacts local and distal budding determinants, revealing additional late domain activities
- PMID: 24257210
- PMCID: PMC3907034
- DOI: 10.1186/1742-4690-10-143
The S40 residue in HIV-1 Gag p6 impacts local and distal budding determinants, revealing additional late domain activities
Abstract
Background: HIV-1 budding is directed primarily by two motifs in Gag p6 designated as late domain-1 and -2 that recruit ESCRT machinery by binding Tsg101 and Alix, respectively, and by poorly characterized determinants in the capsid (CA) domain. Here, we report that a conserved Gag p6 residue, S40, impacts budding mediated by all of these determinants.
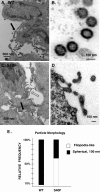
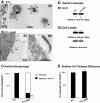
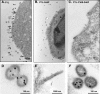
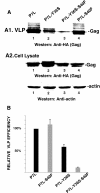
Results: Whereas budding normally results in formation of single spherical particles ~100 nm in diameter and containing a characteristic electron-dense conical core, the substitution of Phe for S40, a change that does not alter the amino acids encoded in the overlapping pol reading frame, resulted in defective CA-SP1 cleavage, formation of strings of tethered particles or filopodia-like membrane protrusions containing Gag, and diminished infectious particle formation. The S40F-mediated release defects were exacerbated when the viral-encoded protease (PR) was inactivated or when L domain-1 function was disrupted or when budding was almost completely obliterated by the disruption of both L domain-1 and -2. S40F mutation also resulted in stronger Gag-Alix interaction, as detected by yeast 2-hybrid assay. Reducing Alix binding by mutational disruption of contact residues restored single particle release, implicating the perturbed Gag-Alix interaction in the aberrant budding events. Interestingly, introduction of S40F partially rescued the negative effects on budding of CA NTD mutations EE75,76AA and P99A, which both prevent membrane curvature and therefore block budding at an early stage.
Conclusions: The results indicate that the S40 residue is a novel determinant of HIV-1 egress that is most likely involved in regulation of a critical assembly event required for budding in the Tsg101-, Alix-, Nedd4- and CA N-terminal domain affected pathways.
Figures



References
-
- Gottlinger HG. The HIV-1 assembly machinery. AIDS. 2001;10(Suppl 5):S13–20. - PubMed
-
- Garrus JE, Von Schwedler UK, Pornillos OW, Morham SG, Zavitz KH, Wang HE, Wettstein DA, Stray KM, Cote M, Rich RL, Myszka DG, Sundquist WI. Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell Dev Biol. 2001;10:55–65. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

