Overlapping and distinct roles of Aspergillus fumigatus UDP-glucose 4-epimerases in galactose metabolism and the synthesis of galactose-containing cell wall polysaccharides
- PMID: 24257745
- PMCID: PMC3894311
- DOI: 10.1074/jbc.M113.522516
Overlapping and distinct roles of Aspergillus fumigatus UDP-glucose 4-epimerases in galactose metabolism and the synthesis of galactose-containing cell wall polysaccharides
Abstract
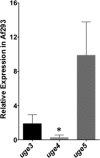
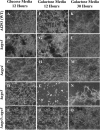
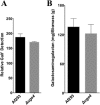
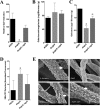
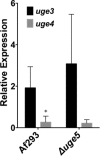
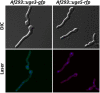
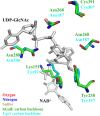
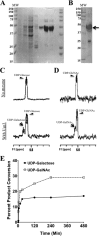
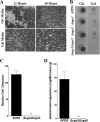
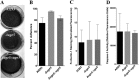
The cell wall of Aspergillus fumigatus contains two galactose-containing polysaccharides, galactomannan and galactosaminogalactan, whose biosynthetic pathways are not well understood. The A. fumigatus genome contains three genes encoding putative UDP-glucose 4-epimerases, uge3, uge4, and uge5. We undertook this study to elucidate the function of these epimerases. We found that uge4 is minimally expressed and is not required for the synthesis of galactose-containing exopolysaccharides or galactose metabolism. Uge5 is the dominant UDP-glucose 4-epimerase in A. fumigatus and is essential for normal growth in galactose-based medium. Uge5 is required for synthesis of the galactofuranose (Galf) component of galactomannan and contributes galactose to the synthesis of galactosaminogalactan. Uge3 can mediate production of both UDP-galactose and UDP-N-acetylgalactosamine (GalNAc) and is required for the production of galactosaminogalactan but not galactomannan. In the absence of Uge5, Uge3 activity is sufficient for growth on galactose and the synthesis of galactosaminogalactan containing lower levels of galactose but not the synthesis of Galf. A double deletion of uge5 and uge3 blocked growth on galactose and synthesis of both Galf and galactosaminogalactan. This study is the first survey of glucose epimerases in A. fumigatus and contributes to our understanding of the role of these enzymes in metabolism and cell wall synthesis.
Keywords: Carbohydrate Biosynthesis; Cell Wall; Galactose Metabolism; Glycobiology; Mycology; Polysaccharide.
Figures

References
-
- Ostrosky-Zeichner L., Casadevall A., Galgiani J. N., Odds F. C., Rex J. H. (2010) An insight into the antifungal pipeline: selected new molecules and beyond. Nat. Rev. Drug Discov. 9, 719–727 - PubMed
-
- Latgé J.-P. (2010) Tasting the fungal cell wall. Cell. Microbiol. 12, 863–872 - PubMed
-
- Latge J. P. (2009) Galactofuranose containing molecules in Aspergillus fumigatus. Med. Mycol. 47, S104–S109 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

