Induction of recombination between diverged sequences in a mammalian genome by a double-strand break
- PMID: 24257896
- PMCID: PMC11113419
- DOI: 10.1007/s00018-013-1520-0
Induction of recombination between diverged sequences in a mammalian genome by a double-strand break
Abstract
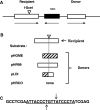
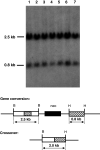
To investigate whether mammalian cells can carry out recombinational double-strand break (DSB) repair between highly diverged sequences, mouse fibroblasts were transfected with DNA substrates that contained a "recipient" thymidine kinase (tk) gene disrupted by the recognition site for endonuclease I-SceI. Substrates also contained a linked "donor" tk gene sequence. Following DSB induction by I-SceI, selection for tk-expressing clones allowed recovery of repair events occurring by nonhomologous end-joining or recombination with the donor sequence. Although recombinational repair was most efficient when donor and recipient shared near-perfect homology, we recovered recombination events between recipient and donor sequences displaying 20 % nucleotide mismatch. Recombination between such imperfectly matched ("homeologous") sequences occurred at a frequency of 1.7 × 10(-7) events per cell and constituted 3 % of the DSB repair events recovered with the pair of homeologous sequences. Additional experiments were done with a substrate containing a donor sequence comprised of a region sharing high homology with the recipient and an adjacent region homeologous to the recipient. Recombinational DSB repair tracts initiating within high homology propagated into homeology in 11 of 112 repair events. These collective results contrasted with our earlier work in which spontaneous recombination (not intentionally induced by a DSB) between homeologous sequences occurred at an undetectable frequency of less than 10(-9) events per cell, and in which events initiating within high homology propagated into adjoining homeology in one of 81 events examined. Our current work suggests that homology requirements for recombination are effectively relaxed in proximity to a DSB in a mammalian genome.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

