Genome-wide association study of extreme longevity in Drosophila melanogaster
- PMID: 24259311
- PMCID: PMC3914684
- DOI: 10.1093/gbe/evt180
Genome-wide association study of extreme longevity in Drosophila melanogaster
Abstract
Human genome-wide association studies (GWAS) of longevity attempt to identify alleles at different frequencies in the extremely old, relative to a younger control sample. Here, we apply a GWAS approach to "synthetic" populations of Drosophila melanogaster derived from a small number of inbred founders. We used next-generation DNA sequencing to estimate allele and haplotype frequencies in the oldest surviving individuals of an age cohort and compared these frequencies with those of randomly sampled individuals from the same cohort. We used this case-control strategy in four independent cohorts and identified eight significantly differentiated regions of the genome potentially harboring genes with relevance for longevity. By modeling the effects of local haplotypes, we have more power to detect regions enriched for longevity genes than marker-based GWAS. Most significant regions occur near chromosome ends or centromeres where recombination is infrequent, consistent with these regions harboring unconditionally deleterious alleles impacting longevity. Genes in regions of normal recombination are enriched for those relevant to immune function and a gene family involved in oxidative stress response. Genetic differentiation between our experimental cohorts is comparable to that between human populations, suggesting in turn that our results may help explain heterogeneous signals in human association studies of extreme longevity when panels have diverse ancestry.
Keywords: GWAS; ageing; synthetic populations.
Figures
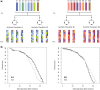
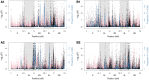
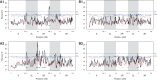
Comment in
-
Highlight: delaying the call of death--genome-wide study of long-lived fruit flies.Genome Biol Evol. 2014 Jan;6(1):34-5. doi: 10.1093/gbe/evt203. Genome Biol Evol. 2014. PMID: 24391161 Free PMC article. No abstract available.
Similar articles
-
Nucleotide diversity inflation as a genome-wide response to experimental lifespan extension in Drosophila melanogaster.BMC Genomics. 2017 Jan 14;18(1):84. doi: 10.1186/s12864-017-3485-0. BMC Genomics. 2017. PMID: 28088192 Free PMC article.
-
Identification of novel genes associated with longevity in Drosophila melanogaster - a computational approach.Aging (Albany NY). 2019 Dec 3;11(23):11244-11267. doi: 10.18632/aging.102527. Epub 2019 Dec 3. Aging (Albany NY). 2019. PMID: 31794428 Free PMC article.
-
Genetic variation and human longevity.Dan Med J. 2012 May;59(5):B4454. Dan Med J. 2012. PMID: 22549493
-
Dissecting the genetics of longevity in Drosophila melanogaster.Fly (Austin). 2009 Jan-Mar;3(1):29-38. doi: 10.4161/fly.3.1.7771. Epub 2009 Jan 6. Fly (Austin). 2009. PMID: 19182541 Review.
-
Genetics of longevity.Exp Gerontol. 1998 Nov-Dec;33(7-8):773-83. doi: 10.1016/s0531-5565(98)00027-8. Exp Gerontol. 1998. PMID: 9951621 Review.
Cited by
-
Diverse biological processes coordinate the transcriptional response to nutritional changes in a Drosophila melanogaster multiparent population.BMC Genomics. 2020 Jan 28;21(1):84. doi: 10.1186/s12864-020-6467-6. BMC Genomics. 2020. PMID: 31992183 Free PMC article.
-
Deleterious mutations show increasing negative effects with age in Drosophila melanogaster.BMC Biol. 2020 Sep 30;18(1):128. doi: 10.1186/s12915-020-00858-5. BMC Biol. 2020. PMID: 32993647 Free PMC article.
-
Genetic analysis of variation in lifespan using a multiparental advanced intercross Drosophila mapping population.BMC Genet. 2016 Aug 2;17:113. doi: 10.1186/s12863-016-0419-9. BMC Genet. 2016. PMID: 27485207 Free PMC article.
-
Life-History Evolution and the Genetics of Fitness Components in Drosophila melanogaster.Genetics. 2020 Jan;214(1):3-48. doi: 10.1534/genetics.119.300160. Genetics. 2020. PMID: 31907300 Free PMC article. Review.
-
Highlight: delaying the call of death--genome-wide study of long-lived fruit flies.Genome Biol Evol. 2014 Jan;6(1):34-5. doi: 10.1093/gbe/evt203. Genome Biol Evol. 2014. PMID: 24391161 Free PMC article. No abstract available.
References
-
- Anselmi CV, et al. Association of the FOXO3A locus with extreme longevity in a southern Italian centenarian study. Rejuvenation Res. 2009;12:95–104. - PubMed
-
- Burkhard DU, Ward PI, Blanckenhorn WU. Using age grading by wing injuries to estimate size-dependent adult survivorship in the field: a case study of the yellow dung fly Scathophaga stercoraria. Ecol Entomol. 2002;27:514–520.
-
- Charlesworth B. Evolution in age-structured populations. Cambridge: Cambridge University Press; 1994.
-
- Deckert-Cruz DJ, Tyler RH, Landmesser JE, Rose MR. Allozymic differentiation in response to laboratory demographic selection of Drosophila melanogaster. Evolution. 1997;51:865–872. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

