The Smad7-Skp2 complex orchestrates Myc stability, impacting on the cytostatic effect of TGF-β
- PMID: 24259667
- PMCID: PMC3889399
- DOI: 10.1242/jcs.136028
The Smad7-Skp2 complex orchestrates Myc stability, impacting on the cytostatic effect of TGF-β
Abstract
In most human cancers the Myc proto-oncogene is highly activated. Dysregulation of Myc oncoprotein contributes to tumorigenesis in numerous tissues and organs. Thus, targeting Myc stability could be a crucial step for cancer therapy. Here we report Smad7 as a key molecule regulating Myc stability and activity by recruiting the F-box protein, Skp2. Ectopic expression of Smad7 downregulated the protein level of Myc without affecting the transcription level, and significantly repressed its transcriptional activity, leading to inhibition of cell proliferation and tumorigenic activity. Furthermore, Smad7 enhanced ubiquitylation of Myc through direct interaction with Myc and recruitment of Skp2. Ablation of Smad7 resulted in less sensitivity to the growth inhibitory effect of TGF-β by inducing stable Myc expression. In conclusion, these findings that Smad7 functions in Myc oncoprotein degradation and enhances the cytostatic effect of TGF-β signaling provide a possible new therapeutic approach for cancer treatment.
Keywords: Myc; Protein stability; Skp2; Smad7; Ubiquitylation.
Figures

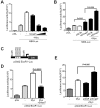

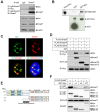


References
-
- Adhikary S., Marinoni F., Hock A., Hulleman E., Popov N., Beier R., Bernard S., Quarto M., Capra M., Goettig S. et al. (2005). The ubiquitin ligase HectH9 regulates transcriptional activation by Myc and is essential for tumor cell proliferation. Cell 123, 409–421 10.1016/j.cell.2005.08.016 - DOI - PubMed
-
- Bahram F., von der Lehr N., Cetinkaya C., Larsson L. G. (2000). c-Myc hot spot mutations in lymphomas result in inefficient ubiquitination and decreased proteasome-mediated turnover. Blood 95, 2104–2110 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

