Evolutionary mix-and-match with MFS transporters II
- PMID: 24259711
- PMCID: PMC3864288
- DOI: 10.1073/pnas.1319754110
Evolutionary mix-and-match with MFS transporters II
Abstract
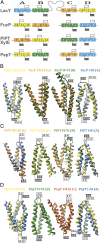
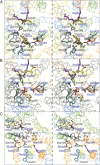
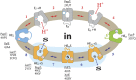
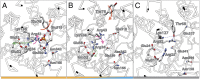
One fundamentally important problem for understanding the mechanism of coupling between substrate and H(+) translocation with secondary active transport proteins is the identification and physical localization of residues involved in substrate and H(+) binding. This information is exceptionally difficult to obtain with the Major Facilitator Superfamily (MFS) because of the broad sequence diversity of the members. The MFS is the largest and most diverse group of transporters, many of which are clinically important, and includes members from all kingdoms of life. A wide range of substrates is transported, in many instances against a concentration gradient by transduction of the energy stored in an H(+) electrochemical gradient using symport mechanisms, which are discussed herein. Crystallographic structures of MFS members indicate that a deep central hydrophilic cavity surrounded by 12 mostly irregular transmembrane helices represents a common structural feature. An inverted triple-helix structural symmetry motif within the N- and C-terminal six-helix bundles suggests that the proteins may have arisen by intragenic multiplication. In the work presented here, the triple-helix motifs are aligned in combinatorial fashion so as to detect functionally homologous positions with known atomic structures of MFS members. Substrate and H(+)-binding sites in symporters that transport substrates, ranging from simple ions like phosphate to more complex peptides or disaccharides, are found to be in similar locations. It also appears likely that there is a homologous ordered kinetic mechanism for the H(+)-coupled MFS symporters.
Keywords: bioenergetics; membrane transport; sequence alignment.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Evolutionary mix-and-match with MFS transporters.Proc Natl Acad Sci U S A. 2013 Apr 9;110(15):5870-4. doi: 10.1073/pnas.1303538110. Epub 2013 Mar 25. Proc Natl Acad Sci U S A. 2013. PMID: 23530251 Free PMC article.
-
Comparative Sequence-Function Analysis of the Major Facilitator Superfamily: The "Mix-and-Match" Method.Methods Enzymol. 2015;557:521-49. doi: 10.1016/bs.mie.2014.12.015. Epub 2015 Mar 24. Methods Enzymol. 2015. PMID: 25950980
-
Integration of evolutionary features for the identification of functionally important residues in major facilitator superfamily transporters.PLoS Comput Biol. 2009 Oct;5(10):e1000522. doi: 10.1371/journal.pcbi.1000522. Epub 2009 Oct 2. PLoS Comput Biol. 2009. PMID: 19798434 Free PMC article.
-
From membrane to molecule to the third amino acid from the left with a membrane transport protein.Q Rev Biophys. 1997 Nov;30(4):333-64. doi: 10.1017/s0033583597003387. Q Rev Biophys. 1997. PMID: 9634651 Review.
-
Energy coupling mechanisms of MFS transporters.Protein Sci. 2015 Oct;24(10):1560-79. doi: 10.1002/pro.2759. Epub 2015 Sep 18. Protein Sci. 2015. PMID: 26234418 Free PMC article. Review.
Cited by
-
pH Regulation of Electrogenic Sugar/H+ Symport in MFS Sugar Permeases.PLoS One. 2016 May 26;11(5):e0156392. doi: 10.1371/journal.pone.0156392. eCollection 2016. PLoS One. 2016. PMID: 27227677 Free PMC article.
-
Variations in exons 11 and 12 of the multi-pest resistance wheat gene Lr34 are independently additive for leaf rust resistance.Front Plant Sci. 2023 Feb 23;13:1061490. doi: 10.3389/fpls.2022.1061490. eCollection 2022. Front Plant Sci. 2023. PMID: 36910459 Free PMC article.
-
Structures and General Transport Mechanisms by the Major Facilitator Superfamily (MFS).Chem Rev. 2021 May 12;121(9):5289-5335. doi: 10.1021/acs.chemrev.0c00983. Epub 2021 Apr 22. Chem Rev. 2021. PMID: 33886296 Free PMC article.
-
Crystal Structures of the Extracellular Domain from PepT1 and PepT2 Provide Novel Insights into Mammalian Peptide Transport.Structure. 2015 Oct 6;23(10):1889-1899. doi: 10.1016/j.str.2015.07.016. Epub 2015 Aug 27. Structure. 2015. PMID: 26320580 Free PMC article.
-
Major Facilitator Superfamily (MFS) evolved without 3-transmembrane segment unit rearrangements.Proc Natl Acad Sci U S A. 2014 Apr 1;111(13):E1162-3. doi: 10.1073/pnas.1400016111. Epub 2014 Feb 24. Proc Natl Acad Sci U S A. 2014. PMID: 24567407 Free PMC article. No abstract available.
References
-
- Saier MH., Jr Families of transmembrane sugar transport proteins. Mol Microbiol. 2000;35(4):699–710. - PubMed
-
- Saier MH, Jr, et al. The major facilitator superfamily. J Mol Microbiol Biotechnol. 1999;1(2):257–279. - PubMed
-
- Mitchell P. Translocations through natural membranes. Adv Enzymol Relat Areas Mol Biol. 1967;29:33–87. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

