Malaria life cycle intensifies both natural selection and random genetic drift
- PMID: 24259712
- PMCID: PMC3864301
- DOI: 10.1073/pnas.1319857110
Malaria life cycle intensifies both natural selection and random genetic drift
Abstract
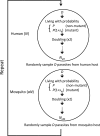
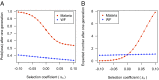
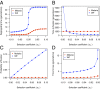
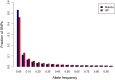
Analysis of genome sequences of 159 isolates of Plasmodium falciparum from Senegal yields an extraordinarily high proportion (26.85%) of protein-coding genes with the ratio of nonsynonymous to synonymous polymorphism greater than one. This proportion is much greater than observed in other organisms. Also unusual is that the site-frequency spectra of synonymous and nonsynonymous polymorphisms are virtually indistinguishable. We hypothesized that the complicated life cycle of malaria parasites might lead to qualitatively different population genetics from that predicted from the classical Wright-Fisher (WF) model, which assumes a single random-mating population with a finite and constant population size in an organism with nonoverlapping generations. This paper summarizes simulation studies of random genetic drift and selection in malaria parasites that take into account their unusual life history. Our results show that random genetic drift in the malaria life cycle is more pronounced than under the WF model. Paradoxically, the efficiency of purifying selection in the malaria life cycle is also greater than under WF, and the relative efficiency of positive selection varies according to conditions. Additionally, the site-frequency spectrum under neutrality is also more skewed toward low-frequency alleles than expected with WF. These results highlight the importance of considering the malaria life cycle when applying existing population genetic tools based on the WF model. The same caveat applies to other species with similarly complex life cycles.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
Publication types
MeSH terms
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

