In silico molecular comparisons of C. elegans and mammalian pharmacology identify distinct targets that regulate feeding
- PMID: 24260022
- PMCID: PMC3833878
- DOI: 10.1371/journal.pbio.1001712
In silico molecular comparisons of C. elegans and mammalian pharmacology identify distinct targets that regulate feeding
Abstract
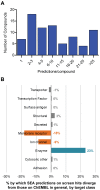
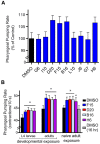
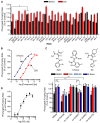
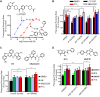
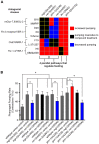
Phenotypic screens can identify molecules that are at once penetrant and active on the integrated circuitry of a whole cell or organism. These advantages are offset by the need to identify the targets underlying the phenotypes. Additionally, logistical considerations limit screening for certain physiological and behavioral phenotypes to organisms such as zebrafish and C. elegans. This further raises the challenge of elucidating whether compound-target relationships found in model organisms are preserved in humans. To address these challenges we searched for compounds that affect feeding behavior in C. elegans and sought to identify their molecular mechanisms of action. Here, we applied predictive chemoinformatics to small molecules previously identified in a C. elegans phenotypic screen likely to be enriched for feeding regulatory compounds. Based on the predictions, 16 of these compounds were tested in vitro against 20 mammalian targets. Of these, nine were active, with affinities ranging from 9 nM to 10 µM. Four of these nine compounds were found to alter feeding. We then verified the in vitro findings in vivo through genetic knockdowns, the use of previously characterized compounds with high affinity for the four targets, and chemical genetic epistasis, which is the effect of combined chemical and genetic perturbations on a phenotype relative to that of each perturbation in isolation. Our findings reveal four previously unrecognized pathways that regulate feeding in C. elegans with strong parallels in mammals. Together, our study addresses three inherent challenges in phenotypic screening: the identification of the molecular targets from a phenotypic screen, the confirmation of the in vivo relevance of these targets, and the evolutionary conservation and relevance of these targets to their human orthologs.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

Comment in
-
From complex behavior to new drugs: chemoinformatics shows the way.PLoS Biol. 2013 Nov;11(11):e1001713. doi: 10.1371/journal.pbio.1001713. Epub 2013 Nov 19. PLoS Biol. 2013. PMID: 24260023 Free PMC article. No abstract available.
Similar articles
-
Effects of genetic mutations and chemical exposures on Caenorhabditis elegans feeding: evaluation of a novel, high-throughput screening assay.PLoS One. 2007 Dec 5;2(12):e1259. doi: 10.1371/journal.pone.0001259. PLoS One. 2007. PMID: 18060055 Free PMC article.
-
A rapid in vivo pipeline to identify small molecule inhibitors of amyloid aggregation.Nat Commun. 2024 Sep 27;15(1):8311. doi: 10.1038/s41467-024-52480-6. Nat Commun. 2024. PMID: 39333123 Free PMC article.
-
Chemical activation of a food deprivation signal extends lifespan.Aging Cell. 2016 Oct;15(5):832-41. doi: 10.1111/acel.12492. Epub 2016 May 24. Aging Cell. 2016. PMID: 27220516 Free PMC article.
-
Serotonergic modulation of feeding behavior in Caenorhabditis elegans and other related nematodes.Neurosci Res. 2020 May;154:9-19. doi: 10.1016/j.neures.2019.04.006. Epub 2019 Apr 24. Neurosci Res. 2020. PMID: 31028772 Review.
-
Neural Regulatory Pathways of Feeding and Fat in Caenorhabditis elegans.Annu Rev Genet. 2015;49:413-38. doi: 10.1146/annurev-genet-120213-092244. Epub 2015 Oct 14. Annu Rev Genet. 2015. PMID: 26473379 Review.
Cited by
-
Integrated Approaches for Genome-wide Interrogation of the Druggable Non-olfactory G Protein-coupled Receptor Superfamily.J Biol Chem. 2015 Aug 7;290(32):19471-7. doi: 10.1074/jbc.R115.654764. Epub 2015 Jun 22. J Biol Chem. 2015. PMID: 26100629 Free PMC article. Review.
-
Ligand Similarity Complements Sequence, Physical Interaction, and Co-Expression for Gene Function Prediction.PLoS One. 2016 Jul 28;11(7):e0160098. doi: 10.1371/journal.pone.0160098. eCollection 2016. PLoS One. 2016. PMID: 27467773 Free PMC article.
-
The steroid hormone ADIOL promotes learning by reducing neural kynurenic acid levels.Genes Dev. 2023 Dec 26;37(21-24):998-1016. doi: 10.1101/gad.350745.123. Genes Dev. 2023. PMID: 38092521 Free PMC article.
-
LITTLE FISH, BIG DATA: ZEBRAFISH AS A MODEL FOR CARDIOVASCULAR AND METABOLIC DISEASE.Physiol Rev. 2017 Jul 1;97(3):889-938. doi: 10.1152/physrev.00038.2016. Physiol Rev. 2017. PMID: 28468832 Free PMC article. Review.
-
Transgenerational inheritance of metabolic disease.Semin Cell Dev Biol. 2015 Jul;43:131-140. doi: 10.1016/j.semcdb.2015.04.007. Epub 2015 Apr 29. Semin Cell Dev Biol. 2015. PMID: 25937492 Free PMC article. Review.
References
-
- Ahlquist RP (1948) A study of the adrenotropic receptors. Am J Physiol 153: 586–600. - PubMed
-
- Black JW, Duncan WA, Durant CJ, Ganellin CR, Parsons EM (1972) Definition and antagonism of histamine H 2 -receptors. Nature 236: 385–390. - PubMed
-
- Gilbert PE, Martin WR (1976) The effects of morphine and nalorphine-like drugs in the nondependent, morphine-dependent and cyclazocine-dependent chronic spinal dog. J Pharmacol Exp Ther 198: 66–82. - PubMed
-
- Richardson BP, Engel G, Donatsch P, Stadler PA (1985) Identification of serotonin M-receptor subtypes and their specific blockade by a new class of drugs. Nature 316: 126–131. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- ES019458/ES/NIEHS NIH HHS/United States
- R44 GM093456/GM/NIGMS NIH HHS/United States
- R21 ES021412/ES/NIEHS NIH HHS/United States
- ES021412-01/ES/NIEHS NIH HHS/United States
- GM93456/GM/NIGMS NIH HHS/United States
- GM081863/GM/NIGMS NIH HHS/United States
- R21 NS082856/NS/NINDS NIH HHS/United States
- U01 ES019458/ES/NIEHS NIH HHS/United States
- R43 GM093456/GM/NIGMS NIH HHS/United States
- R01 GM071896/GM/NIGMS NIH HHS/United States
- GM71896/GM/NIGMS NIH HHS/United States
- R01 GM081863/GM/NIGMS NIH HHS/United States
- R43 GM097912/GM/NIGMS NIH HHS/United States
- GM97912/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

