The contributions of HIF-target genes to tumor growth in RCC
- PMID: 24260413
- PMCID: PMC3832366
- DOI: 10.1371/journal.pone.0080544
The contributions of HIF-target genes to tumor growth in RCC
Abstract
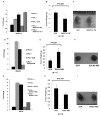
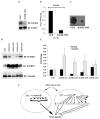
Somatic mutations or loss of expression of tumor suppressor VHL happen in the vast majority of clear cell Renal Cell Carcinoma, and it's causal for kidney cancer development. Without VHL, constitutively active transcription factor HIF is strongly oncogenic and is essential for tumor growth. However, the contribution of individual HIF-responsive genes to tumor growth is not well understood. In this study we examined the contribution of important HIF-responsive genes such as VEGF, CCND1, ANGPTL4, EGLN3, ENO2, GLUT1 and IGFBP3 to tumor growth in a xenograft model using immune-compromised nude mice. We found that the suppression of VEGF or CCND1 impaired tumor growth, suggesting that they are tumor-promoting genes. We further discovered that the lack of ANGPTL4, EGLN3 or ENO2 expression did not change tumor growth. Surprisingly, depletion of GLUT1 or IGFBP3 significantly increased tumor growth, suggesting that they have tumor-inhibitory functions. Depletion of IGFBP3 did not lead to obvious activation of IGFIR. Unexpectedly, the depletion of IGFIR protein led to significant increase of IGFBP3 at both the protein and mRNA levels. Concomitantly, the tumor growth was greatly impaired, suggesting that IGFBP3 might suppress tumor growth in an IGFIR-independent manner. In summary, although the overall transcriptional activity of HIF is strongly tumor-promoting, the expression of each individual HIF-responsive gene could either enhance, reduce or do nothing to the kidney cancer tumor growth.
Conflict of interest statement
Figures




Similar articles
-
The von Hippel-Lindau tumor suppressor protein regulates gene expression and tumor growth through histone demethylase JARID1C.Oncogene. 2012 Feb 9;31(6):776-86. doi: 10.1038/onc.2011.266. Epub 2011 Jul 4. Oncogene. 2012. PMID: 21725364 Free PMC article.
-
Prolyl hydroxylase 2 dependent and Von-Hippel-Lindau independent degradation of Hypoxia-inducible factor 1 and 2 alpha by selenium in clear cell renal cell carcinoma leads to tumor growth inhibition.BMC Cancer. 2012 Jul 17;12:293. doi: 10.1186/1471-2407-12-293. BMC Cancer. 2012. PMID: 22804960 Free PMC article.
-
Identification of cyclin D1 and other novel targets for the von Hippel-Lindau tumor suppressor gene by expression array analysis and investigation of cyclin D1 genotype as a modifier in von Hippel-Lindau disease.Cancer Res. 2002 Jul 1;62(13):3803-11. Cancer Res. 2002. PMID: 12097293
-
The VHL/HIF axis in clear cell renal carcinoma.Semin Cancer Biol. 2013 Feb;23(1):18-25. doi: 10.1016/j.semcancer.2012.06.001. Epub 2012 Jun 13. Semin Cancer Biol. 2013. PMID: 22705278 Free PMC article. Review.
-
The von Hippel-Lindau tumor suppressor protein and clear cell renal carcinoma.Clin Cancer Res. 2007 Jan 15;13(2 Pt 2):680s-684s. doi: 10.1158/1078-0432.CCR-06-1865. Clin Cancer Res. 2007. PMID: 17255293 Review.
Cited by
-
von-Hippel Lindau and Hypoxia-Inducible Factor at the Center of Renal Cell Carcinoma Biology.Hematol Oncol Clin North Am. 2023 Oct;37(5):809-825. doi: 10.1016/j.hoc.2023.04.011. Epub 2023 Jun 1. Hematol Oncol Clin North Am. 2023. PMID: 37270382 Free PMC article. Review.
-
Single cell atlas of kidney cancer endothelial cells reveals distinct expression profiles and phenotypes.BJC Rep. 2024 Mar 14;2(1):23. doi: 10.1038/s44276-024-00047-9. BJC Rep. 2024. PMID: 39516665 Free PMC article.
-
Bioinformatic identification of key genes and analysis of prognostic values in clear cell renal cell carcinoma.Oncol Lett. 2018 Aug;16(2):1747-1757. doi: 10.3892/ol.2018.8842. Epub 2018 May 30. Oncol Lett. 2018. PMID: 30008862 Free PMC article.
-
Dihydroartemisinin inhibits glucose uptake and cooperates with glycolysis inhibitor to induce apoptosis in non-small cell lung carcinoma cells.PLoS One. 2015 Mar 23;10(3):e0120426. doi: 10.1371/journal.pone.0120426. eCollection 2015. PLoS One. 2015. PMID: 25799586 Free PMC article.
-
Efficient test for nonlinear dependence of two continuous variables.BMC Bioinformatics. 2015 Aug 19;16:260. doi: 10.1186/s12859-015-0697-7. BMC Bioinformatics. 2015. PMID: 26283601 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

