Novel virus discovery and genome reconstruction from field RNA samples reveals highly divergent viruses in dipteran hosts
- PMID: 24260463
- PMCID: PMC3832450
- DOI: 10.1371/journal.pone.0080720
Novel virus discovery and genome reconstruction from field RNA samples reveals highly divergent viruses in dipteran hosts
Abstract
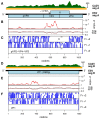
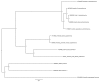
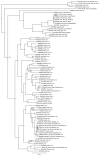
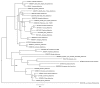
We investigated whether small RNA (sRNA) sequenced from field-collected mosquitoes and chironomids (Diptera) can be used as a proxy signature of viral prevalence within a range of species and viral groups, using sRNAs sequenced from wild-caught specimens, to inform total RNA deep sequencing of samples of particular interest. Using this strategy, we sequenced from adult Anopheles maculipennis s.l. mosquitoes the apparently nearly complete genome of one previously undescribed virus related to chronic bee paralysis virus, and, from a pool of Ochlerotatus caspius and Oc. detritus mosquitoes, a nearly complete entomobirnavirus genome. We also reconstructed long sequences (1503-6557 nt) related to at least nine other viruses. Crucially, several of the sequences detected were reconstructed from host organisms highly divergent from those in which related viruses have been previously isolated or discovered. It is clear that viral transmission and maintenance cycles in nature are likely to be significantly more complex and taxonomically diverse than previously expected.
Conflict of interest statement
Figures









References
-
- Sang RC, Gichogo A, Gachoya J, Dunster MD, Ofula V et al. (2003) Isolation of a new flavivirus related to cell fusing agent virus (CFAV) from field-collected flood-water Aedes mosquitoes sampled from a dambo in central Kenya. Arch Virol 148: 1085-1093. doi:10.1007/s00705-003-0018-8. PubMed: 12756616. - DOI - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

