Multiple transcripts from a 3'-UTR reporter vary in sensitivity to nonsense-mediated mRNA decay in Saccharomyces cerevisiae
- PMID: 24260526
- PMCID: PMC3832414
- DOI: 10.1371/journal.pone.0080981
Multiple transcripts from a 3'-UTR reporter vary in sensitivity to nonsense-mediated mRNA decay in Saccharomyces cerevisiae
Abstract
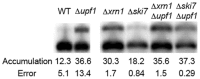
Nonsense-mediated mRNA decay (NMD) causes accelerated transcript degradation when a premature translation termination codon disrupts the open reading frame (ORF). Although endogenous transcripts that have uninterrupted ORFs are typically insensitive to NMD, some can nonetheless become prone to NMD when translation terminates at out-of-frame premature stop codons. This occurs when introns containing stop codons fail to be spliced, when translation of an upstream ORF (uORF) terminates in the 5'-untranslated region (5'-UTR) or the coding region, or when the 5'-proximal AUG initiation codon is bypassed and translation initiates at a downstream out-of-frame AUG followed by a stop codon. Some 3'-untranslated regions (3'-UTRs) are also known to trigger NMD, but the mechanism is less well understood. To further study the role of 3'-UTRs in NMD, a reporter system was designed to examine 3'-UTRs from candidate genes known to produce NMD-sensitive transcripts. Out of eight that were tested, the 3'-UTRs from MSH4 and SPO16 caused NMD-dependent mRNA destabilization. Both endogenous genes produce multiple transcripts that differ in length at the 3' end. Detailed studies revealed that the longest of six reporter MSH4-3'-UTR transcripts was NMD-sensitive but five shorter transcripts were insensitive. NMD-dependent degradation of the long transcript required Xrn1, which degrades mRNA from the 5' end. Sensitivity to NMD was not associated with extensive translational read-through past the normal stop codon. To our knowledge, this is the first example where multiple transcripts containing the same ORF are differentially sensitive to NMD in Saccharomyces cerevisiae. The results provide a proof of principle that long 3'-UTRs can trigger NMD, which suggests a potential link between errors in transcription termination or processing and mRNA decay.
Conflict of interest statement
Figures




Similar articles
-
Long open reading frame transcripts escape nonsense-mediated mRNA decay in yeast.Cell Rep. 2014 Feb 27;6(4):593-8. doi: 10.1016/j.celrep.2014.01.025. Epub 2014 Feb 13. Cell Rep. 2014. PMID: 24529707
-
Nonsense-mediated mRNA decay of metal-binding activator MAC1 is dependent on copper levels and 3'-UTR length in Saccharomyces cerevisiae.Curr Genet. 2024 May 6;70(1):5. doi: 10.1007/s00294-024-01291-9. Curr Genet. 2024. PMID: 38709348
-
The upstream open reading frame of the Arabidopsis AtMHX gene has a strong impact on transcript accumulation through the nonsense-mediated mRNA decay pathway.Plant J. 2009 Dec;60(6):1031-42. doi: 10.1111/j.1365-313X.2009.04021.x. Plant J. 2009. PMID: 19754518
-
NMD monitors translational fidelity 24/7.Curr Genet. 2017 Dec;63(6):1007-1010. doi: 10.1007/s00294-017-0709-4. Epub 2017 May 23. Curr Genet. 2017. PMID: 28536849 Free PMC article. Review.
-
Nonsense in the testis: multiple roles for nonsense-mediated decay revealed in male reproduction.Biol Reprod. 2017 May 1;96(5):939-947. doi: 10.1093/biolre/iox033. Biol Reprod. 2017. PMID: 28444146 Free PMC article. Review.
Cited by
-
A Brief Journey into the History of and Future Sources and Uses of Fatty Acids.Front Nutr. 2021 Jul 20;8:570401. doi: 10.3389/fnut.2021.570401. eCollection 2021. Front Nutr. 2021. PMID: 34355007 Free PMC article. Review.
-
The expression of Rpb10, a small subunit common to RNA polymerases, is modulated by the R3H domain-containing Rbs1 protein and the Upf1 helicase.Nucleic Acids Res. 2020 Dec 2;48(21):12252-12268. doi: 10.1093/nar/gkaa1069. Nucleic Acids Res. 2020. PMID: 33231687 Free PMC article.
-
Overlapping open reading frames strongly reduce human and yeast STN1 gene expression and affect telomere function.PLoS Genet. 2018 Aug 1;14(8):e1007523. doi: 10.1371/journal.pgen.1007523. eCollection 2018 Aug. PLoS Genet. 2018. PMID: 30067734 Free PMC article.
-
NMD: At the crossroads between translation termination and ribosome recycling.Biochimie. 2015 Jul;114:2-9. doi: 10.1016/j.biochi.2014.10.027. Epub 2014 Nov 13. Biochimie. 2015. PMID: 25446649 Free PMC article. Review.
-
Production of Fatty Acid-derived valuable chemicals in synthetic microbes.Front Bioeng Biotechnol. 2014 Dec 23;2:78. doi: 10.3389/fbioe.2014.00078. eCollection 2014. Front Bioeng Biotechnol. 2014. PMID: 25566540 Free PMC article. Review.
References
-
- Culbertson MR, Leeds PF (2003) Looking at mRNA decay pathways through the window of molecular evolution. Curr Opin Genet Dev 13: 207–214. - PubMed
-
- Leeds P, Peltz SW, Jacobson A, Culbertson MR (1991) The product of the yeast UPF1 gene is required for rapid turnover of mRNAs containing a premature translational termination codon. Genes Dev 5: 2303–2314. - PubMed
-
- Chamieh H, Ballut L, Bonneau F, Le Hir H (2008) NMD factors UPF2 and UPF3 bridge UPF1 to the exon junction complex and stimulate its RNA helicase activity. Nat Struct Mol Biol 15: 85–93. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

