Reconstruction of the ancestral marsupial karyotype from comparative gene maps
- PMID: 24261750
- PMCID: PMC4222502
- DOI: 10.1186/1471-2148-13-258
Reconstruction of the ancestral marsupial karyotype from comparative gene maps
Abstract
Background: The increasing number of assembled mammalian genomes makes it possible to compare genome organisation across mammalian lineages and reconstruct chromosomes of the ancestral marsupial and therian (marsupial and eutherian) mammals. However, the reconstruction of ancestral genomes requires genome assemblies to be anchored to chromosomes. The recently sequenced tammar wallaby (Macropus eugenii) genome was assembled into over 300,000 contigs. We previously devised an efficient strategy for mapping large evolutionarily conserved blocks in non-model mammals, and applied this to determine the arrangement of conserved blocks on all wallaby chromosomes, thereby permitting comparative maps to be constructed and resolve the long debated issue between a 2n = 14 and 2n = 22 ancestral marsupial karyotype.
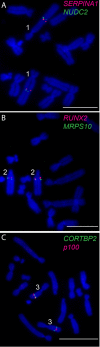
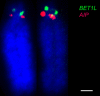
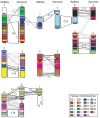
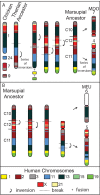
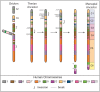
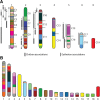
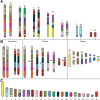
Results: We identified large blocks of genes conserved between human and opossum, and mapped genes corresponding to the ends of these blocks by fluorescence in situ hybridization (FISH). A total of 242 genes was assigned to wallaby chromosomes in the present study, bringing the total number of genes mapped to 554 and making it the most densely cytogenetically mapped marsupial genome. We used these gene assignments to construct comparative maps between wallaby and opossum, which uncovered many intrachromosomal rearrangements, particularly for genes found on wallaby chromosomes X and 3. Expanding comparisons to include chicken and human permitted the putative ancestral marsupial (2n = 14) and therian mammal (2n = 19) karyotypes to be reconstructed.
Conclusions: Our physical mapping data for the tammar wallaby has uncovered the events shaping marsupial genomes and enabled us to predict the ancestral marsupial karyotype, supporting a 2n = 14 ancestor. Futhermore, our predicted therian ancestral karyotype has helped to understand the evolution of the ancestral eutherian genome.
Figures


Similar articles
-
Chromosome evolution in kangaroos (Marsupialia: Macropodidae): cross species chromosome painting between the tammar wallaby and rock wallaby spp. with the 2n = 22 ancestral macropodid karyotype.Genome. 1999 Jun;42(3):525-30. doi: 10.1139/g98-159. Genome. 1999. PMID: 10382300
-
Physical map of two tammar wallaby chromosomes: a strategy for mapping in non-model mammals.Chromosome Res. 2008;16(8):1159-75. doi: 10.1007/s10577-008-1266-y. Epub 2008 Nov 8. Chromosome Res. 2008. PMID: 18987984
-
A second-generation anchored genetic linkage map of the tammar wallaby (Macropus eugenii).BMC Genet. 2011 Aug 19;12:72. doi: 10.1186/1471-2156-12-72. BMC Genet. 2011. PMID: 21854616 Free PMC article.
-
Marsupials in the age of genomics.Annu Rev Genomics Hum Genet. 2013;14:393-420. doi: 10.1146/annurev-genom-091212-153452. Epub 2013 Jul 3. Annu Rev Genomics Hum Genet. 2013. PMID: 23834319 Review.
-
The evolution of marsupial and monotreme chromosomes.Cytogenet Genome Res. 2012;137(2-4):113-29. doi: 10.1159/000339433. Epub 2012 Jul 6. Cytogenet Genome Res. 2012. PMID: 22777195 Review.
Cited by
-
The methylation and telomere landscape in two families of marsupials with different rates of chromosome evolution.Chromosome Res. 2018 Dec;26(4):317-332. doi: 10.1007/s10577-018-9593-0. Epub 2018 Dec 12. Chromosome Res. 2018. PMID: 30539406
-
Global DNA Methylation patterns on marsupial and devil facial tumour chromosomes.Mol Cytogenet. 2015 Oct 1;8:74. doi: 10.1186/s13039-015-0176-x. eCollection 2015. Mol Cytogenet. 2015. PMID: 26435750 Free PMC article.
-
Development of the pulmonary vasculature in the gray short-tailed opossum (Monodelphis domestica)-3D reconstruction by microcomputed tomography.Anat Rec (Hoboken). 2025 Apr;308(4):1144-1163. doi: 10.1002/ar.25542. Epub 2024 Jul 12. Anat Rec (Hoboken). 2025. PMID: 38993078 Free PMC article.
-
Adaptation and conservation insights from the koala genome.Nat Genet. 2018 Aug;50(8):1102-1111. doi: 10.1038/s41588-018-0153-5. Epub 2018 Jul 2. Nat Genet. 2018. PMID: 29967444 Free PMC article.
-
Evolution of the ancestral mammalian karyotype and syntenic regions.Proc Natl Acad Sci U S A. 2022 Oct 4;119(40):e2209139119. doi: 10.1073/pnas.2209139119. Epub 2022 Sep 26. Proc Natl Acad Sci U S A. 2022. PMID: 36161960 Free PMC article.
References
-
- Beck RMD. A dated phylogeny of marsupials using a molecular supermatrix and multiple fossil constraints. J Mammal. 2008;89:175–189. doi: 10.1644/06-MAMM-A-437.1. - DOI
-
- Meredith RW, Westerman M, Case JA, Springer MS. A Phylogeny and timescale for marsupial evolution based on sequences for five nuclear genes. J Mamm Evol. 2008;15:1–36. doi: 10.1007/s10914-007-9062-6. - DOI
-
- Hayman DL. Marsupial cytogenetics. Aust J Zool. 1990;37:331–349.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

