Genome wide gene-expression analysis of facultative reproductive diapause in the two-spotted spider mite Tetranychus urticae
- PMID: 24261877
- PMCID: PMC4046741
- DOI: 10.1186/1471-2164-14-815
Genome wide gene-expression analysis of facultative reproductive diapause in the two-spotted spider mite Tetranychus urticae
Abstract
Background: Diapause or developmental arrest, is one of the major adaptations that allows mites and insects to survive unfavorable conditions. Diapause evokes a number of physiological, morphological and molecular modifications. In general, diapause is characterized by a suppression of the metabolism, change in behavior, increased stress tolerance and often by the synthesis of cryoprotectants. At the molecular level, diapause is less studied but characterized by a complex and regulated change in gene-expression. The spider mite Tetranychus urticae is a serious polyphagous pest that exhibits a reproductive facultative diapause, which allows it to survive winter conditions. Diapausing mites turn deeply orange in color, stop feeding and do not lay eggs.
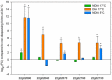
Results: We investigated essential physiological processes in diapausing mites by studying genome-wide expression changes, using a custom built microarray. Analysis of this dataset showed that a remarkable number, 11% of the total number of predicted T. urticae genes, were differentially expressed. Gene Ontology analysis revealed that many metabolic pathways were affected in diapausing females. Genes related to digestion and detoxification, cryoprotection, carotenoid synthesis and the organization of the cytoskeleton were profoundly influenced by the state of diapause. Furthermore, we identified and analyzed an unique class of putative antifreeze proteins that were highly upregulated in diapausing females. We also further confirmed the involvement of horizontally transferred carotenoid synthesis genes in diapause and different color morphs of T. urticae.
Conclusions: This study offers the first in-depth analysis of genome-wide gene-expression patterns related to diapause in a member of the Chelicerata, and further adds to our understanding of the overall strategies of diapause in arthropods.
Figures





References
-
- Denlinger D. Regulation of diapause. Annu Rev Entomol. 2002;47:93–122. - PubMed
-
- Denlinger D. Induction and termination of pupal diapause in Sarcophaga (Diptera: Sarcophagidae) Biol Bull. 1972;142:11–24.
-
- Veerman A. Diapause. In: Sabelis MW, Helle W, editors. Spider mites their Biol Nat enemies Control. Amsterdam: Elsevier; 1985. pp. 279–317.
-
- Hahn DA, Denlinger DL. Meeting the energetic demands of insect diapause: nutrient storage and utilization. J Insect Physiol. 2007;53:760–773. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

