Drugs derived from phage display: from candidate identification to clinical practice
- PMID: 24262785
- PMCID: PMC3929457
- DOI: 10.4161/mabs.27240
Drugs derived from phage display: from candidate identification to clinical practice
Abstract
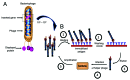
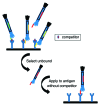
Phage display, one of today’s fundamental drug discovery technologies, allows identification of a broad range of biological drugs, including peptides, antibodies and other proteins, with the ability to tailor critical characteristics such as potency, specificity and cross-species binding. Further, unlike in vivo technologies, generating phage display-derived antibodies is not restricted by immunological tolerance. Although more than 20 phage display-derived antibody and peptides are currently in late-stage clinical trials or approved, there is little literature addressing the specific challenges and successes in the clinical development of phage-derived drugs. This review uses case studies, from candidate identification through clinical development, to illustrate the utility of phage display as a drug discovery tool, and offers a perspective for future developments of phage display technology.
Figures
References
-
- Buckler DR, Schofield D, Sexton DJ, Lowe D, Vaughan TJ. Selection and Screening of Antibody Phage Display Libraries. In: Wood CR, ed. Antibody Drug Discovery. London: Imperial College Press, 2012.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
