Evidence for activation of the unfolded protein response in collagen IV nephropathies
- PMID: 24262798
- PMCID: PMC3904556
- DOI: 10.1681/ASN.2012121217
Evidence for activation of the unfolded protein response in collagen IV nephropathies
Abstract
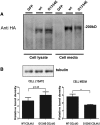
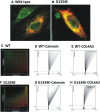
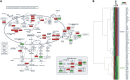
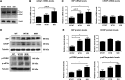
Thin-basement-membrane nephropathy (TBMN) and Alport syndrome (AS) are progressive collagen IV nephropathies caused by mutations in COL4A3/A4/A5 genes. These nephropathies invariably present with microscopic hematuria and frequently progress to proteinuria and CKD or ESRD during long-term follow-up. Nonetheless, the exact molecular mechanisms by which these mutations exert their deleterious effects on the glomerulus remain elusive. We hypothesized that defective trafficking of the COL4A3 chain causes a strong intracellular effect on the cell responsible for COL4A3 expression, the podocyte. To this end, we overexpressed normal and mutant COL4A3 chains (G1334E mutation) in human undifferentiated podocytes and tested their effects in various intracellular pathways using a microarray approach. COL4A3 overexpression in the podocyte caused chain retention in the endoplasmic reticulum (ER) that was associated with activation of unfolded protein response (UPR)-related markers of ER stress. Notably, the overexpression of normal or mutant COL4A3 chains differentially activated the UPR pathway. Similar results were observed in a novel knockin mouse carrying the Col4a3-G1332E mutation, which produced a phenotype consistent with AS, and in biopsy specimens from patients with TBMN carrying a heterozygous COL4A3-G1334E mutation. These results suggest that ER stress arising from defective localization of collagen IV chains in human podocytes contributes to the pathogenesis of TBMN and AS through activation of the UPR, a finding that may pave the way for novel therapeutic interventions for a variety of collagenopathies.
Figures







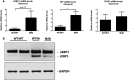
References
-
- Haas M: Alport syndrome and thin glomerular basement membrane nephropathy: a practical approach to diagnosis. Arch Pathol Lab Med 133: 224–232., 2009 - PubMed
-
- Thorner PS: Alport syndrome and thin basement membrane nephropathy. Nephron Clin Pract 106: c82–c88, 2007 - PubMed
-
- Temme J, Peters F, Lange K, Pirson Y, Heidet L, Torra R, Grunfeld JP, Weber M, Licht C, Müller GA, Gross O: Incidence of renal failure and nephroprotection by RAAS inhibition in heterozygous carriers of X-chromosomal and autosomal recessive Alport mutations. Kidney Int 81: 779–783, 2012 - PubMed
-
- Voskarides K, Damianou L, Neocleous V, Zouvani I, Christodoulidou S, Hadjiconstantinou V, Ioannou K, Athanasiou Y, Patsias C, Alexopoulos E, Pierides A, Kyriacou K, Deltas C: COL4A3/COL4A4 mutations producing focal segmental glomerulosclerosis and renal failure in thin basement membrane nephropathy. J Am Soc Nephrol 18: 3004–3016, 2007 - PubMed
-
- Parkin JD, San Antonio JD, Pedchenko V, Hudson B, Jensen ST, Savige J: Mapping structural landmarks, ligand binding sites, and missense mutations to the collagen IV heterotrimers predicts major functional domains, novel interactions, and variation in phenotypes in inherited diseases affecting basement membranes. Hum Mutat 32: 127–143, 2011 - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

