ArchDB 2014: structural classification of loops in proteins
- PMID: 24265221
- PMCID: PMC3964960
- DOI: 10.1093/nar/gkt1189
ArchDB 2014: structural classification of loops in proteins
Abstract
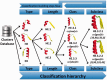
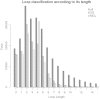
The function of a protein is determined by its three-dimensional structure, which is formed by regular (i.e. β-strands and α-helices) and non-periodic structural units such as loops. Compared to regular structural elements, non-periodic, non-repetitive conformational units enclose a much higher degree of variability--raising difficulties in the identification of regularities, and yet represent an important part of the structure of a protein. Indeed, loops often play a pivotal role in the function of a protein and different aspects of protein folding and dynamics. Therefore, the structural classification of protein loops is an important subject with clear applications in homology modelling, protein structure prediction, protein design (e.g. enzyme design and catalytic loops) and function prediction. ArchDB, the database presented here (freely available at http://sbi.imim.es/archdb), represents such a resource and has been an important asset for the scientific community throughout the years. In this article, we present a completely reworked and updated version of ArchDB. The new version of ArchDB features a novel, fast and user-friendly web-based interface, and a novel graph-based, computationally efficient, clustering algorithm. The current version of ArchDB classifies 149,134 loops in 5739 classes and 9608 subclasses.
Figures

References
-
- Garcia-Garcia J, Bonet J, Guney E, Fornes O, Planas-Iglesias J, Oliva B. Networks of protein–protein interactions: from uncertainty to molecular details. Mol. Inform. 2012;31:342–362. - PubMed
-
- Lee D, Redfern O, Orengo C. Predicting protein function from sequence and structure. Nat. Rev. Mol. Cell Biol. 2007;8:995–1005. - PubMed
-
- Mosca R, Pons T, Ceol A, Valencia A, Aloy P. Towards a detailed atlas of protein-protein interactions. Curr. Opin. Struct. Biol. 2013;23:929–940. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

