Ovarian cancer: genomic analysis
- PMID: 24265410
- PMCID: PMC3836569
- DOI: 10.1093/annonc/mdt462
Ovarian cancer: genomic analysis
Abstract
Objectives: Despite improvements in the management of ovarian cancer patients over the last 30 years, there has been only a minimal improvement in overall survival. While targeted therapeutic approaches for the treatment of cancer have evolved, major challenges in ovarian cancer research persist, including the identification of predictive biomarkers with clinical relevance, so that empirical drug selection can be avoided. In this article, we review published genomic analysis studies including data generated in our laboratory and how they have been incorporated into modern clinical trials in a rational and effective way.
Methods: Multiple published genomic analysis studies were collected for review and discussion with emphasis on their potential clinical applicability.
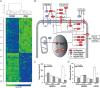
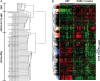
Results: Genomic analysis has been shown to be a powerful tool to identify dysregulated genes, aberrantly activated pathways and to uncover uniqueness of subclasses of ovarian tumors. The application of this technology has provided a solid molecular basis for different clinical behaviors associated with tumor histology and grade. Genomic signatures have been obtained to predict clinical end points for patients with cancer, including response rates, progression-free survival, and overall survival. In addition, genomic analysis has provided opportunities to identify biomarkers, which either result in a modification of existing clinical management or to stratification of patients to novel therapeutic approaches designed as clinical trials.
Conclusions: Genomic analyses have accelerated the identification of relevant biomarkers and extended our understanding of the molecular biology of ovarian cancer. This in turn, will hopefully lead to a paradigm shift from empirical, uniform treatment to a more rational, personalized treatment of ovarian cancers. However, validation of potential biomarkers on both the statistical and biological levels is needed to confirm they are of clinical relevance, in order to increase the likelihood that the desired outcome can be predicted and achieved.
Keywords: cancer; clinical trials; genomics; ovarian.
Figures


References
-
- Colombo N, Peiretti M, Castiglione M. Non-epithelial ovarian cancer: ESMO clinical recommendations for diagnosis, treatment and follow-up. Ann Oncol. 2009;20(Suppl 4):24–26. - PubMed
-
- del Carmen MG, Birrer M, Schorge JO. Carcinosarcoma of the ovary: a review of the literature. Gynecol Oncol. 2012;125:271–277. - PubMed
-
- Bristow RE, Tomacruz RS, Armstrong DK, et al. Survival effect of maximal cytoreductive surgery for advanced ovarian carcinoma during the platinum era: a meta-analysis. J Clin Oncol. 2002;20:1248–1259. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

