PHDcleav: a SVM based method for predicting human Dicer cleavage sites using sequence and secondary structure of miRNA precursors
- PMID: 24267009
- PMCID: PMC3851333
- DOI: 10.1186/1471-2105-14-S14-S9
PHDcleav: a SVM based method for predicting human Dicer cleavage sites using sequence and secondary structure of miRNA precursors
Abstract
Background: Dicer, an RNase III enzyme, plays a vital role in the processing of pre-miRNAs for generating the miRNAs. The structural and sequence features on pre-miRNA which can facilitate position and efficiency of cleavage are not well known. A precise cleavage by Dicer is crucial because an inaccurate processing can produce miRNA with different seed regions which can alter the repertoire of target genes.
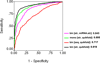
Results: In this study, a novel method has been developed to predict Dicer cleavage sites on pre-miRNAs using Support Vector Machine. We used the dataset of experimentally validated human miRNA hairpins from miRBase, and extracted fourteen nucleotides around Dicer cleavage sites. We developed number of models using various types of features and achieved maximum accuracy of 66% using binary profile of nucleotide sequence taken from 5p arm of hairpin. The prediction performance of Dicer cleavage site improved significantly from 66% to 86% when we integrated secondary structure information. This indicates that secondary structure plays an important role in the selection of cleavage site. All models were trained and tested on 555 experimentally validated cleavage sites and evaluated using 5-fold cross validation technique. In addition, the performance was also evaluated on an independent testing dataset that achieved an accuracy of ~82%.
Conclusion: Based on this study, we developed a webserver PHDcleav (http://www.imtech.res.in/raghava/phdcleav/) to predict Dicer cleavage sites in pre-miRNA. This tool can be used to investigate functional consequences of genetic variations/SNPs in miRNA on Dicer cleavage site, and gene silencing. Moreover, it would also be useful in the discovery of miRNAs in human genome and design of Dicer specific pre-miRNAs for potent gene silencing.
Figures

References
-
- Yang WJ, Yang DD, Na S, Sandusky GE, Zhang Q, Zhao G. Dicer is required for embryonic angiogenesis during mouse development. J Biol Chem. 2005;14(10):9330–9335. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

