The chromatin modification by SUMO-2/3 but not SUMO-1 prevents the epigenetic activation of key immune-related genes during Kaposi's sarcoma associated herpesvirus reactivation
- PMID: 24267727
- PMCID: PMC4046822
- DOI: 10.1186/1471-2164-14-824
The chromatin modification by SUMO-2/3 but not SUMO-1 prevents the epigenetic activation of key immune-related genes during Kaposi's sarcoma associated herpesvirus reactivation
Abstract
Background: SUMOylation, as part of the epigenetic regulation of transcription, has been intensively studied in lower eukaryotes that contain only a single SUMO protein; however, the functions of SUMOylation during mammalian epigenetic transcriptional regulation are largely uncharacterized. Mammals express three major SUMO paralogues: SUMO-1, SUMO-2, and SUMO-3 (normally referred to as SUMO-1 and SUMO-2/3). Herpesviruses, including Kaposi's sarcoma associated herpesvirus (KSHV), seem to have evolved mechanisms that directly or indirectly modulate the SUMO machinery in order to evade host immune surveillance, thus advancing their survival. Interestingly, KSHV encodes a SUMO E3 ligase, K-bZIP, with specificity toward SUMO-2/3 and is an excellent model for investigating the global functional differences between SUMO paralogues.
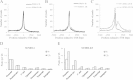
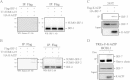
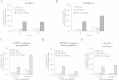
Results: We investigated the effect of experimental herpesvirus reactivation in a KSHV infected B lymphoma cell line on genomic SUMO-1 and SUMO-2/3 binding profiles together with the potential role of chromatin SUMOylation in transcription regulation. This was carried out via high-throughput sequencing analysis. Interestingly, chromatin immunoprecipitation sequencing (ChIP-seq) experiments showed that KSHV reactivation is accompanied by a significant increase in SUMO-2/3 modification around promoter regions, but SUMO-1 enrichment was absent. Expression profiling revealed that the SUMO-2/3 targeted genes are primarily highly transcribed genes that show no expression changes during viral reactivation. Gene ontology analysis further showed that these genes are involved in cellular immune responses and cytokine signaling. High-throughput annotation of SUMO occupancy of transcription factor binding sites (TFBS) pinpointed the presence of three master regulators of immune responses, IRF-1, IRF-2, and IRF-7, as potential SUMO-2/3 targeted transcriptional factors after KSHV reactivation.
Conclusion: Our study is the first to identify differential genome-wide SUMO modifications between SUMO paralogues during herpesvirus reactivation. Our findings indicate that SUMO-2/3 modification near protein-coding gene promoters occurs in order to maintain host immune-related gene unaltered during viral reactivation.
Figures







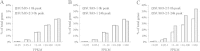


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

