Systematic classification of non-coding RNAs by epigenomic similarity
- PMID: 24267974
- PMCID: PMC3851203
- DOI: 10.1186/1471-2105-14-S14-S2
Systematic classification of non-coding RNAs by epigenomic similarity
Abstract
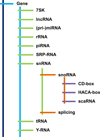
Background: Even though only 1.5% of the human genome is translated into proteins, recent reports indicate that most of it is transcribed into non-coding RNAs (ncRNAs), which are becoming the subject of increased scientific interest. We hypothesized that examining how different classes of ncRNAs co-localized with annotated epigenomic elements could help understand the functions, regulatory mechanisms, and relationships among ncRNA families.
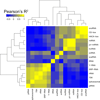
Results: We examined 15 different ncRNA classes for statistically significant genomic co-localizations with cell type-specific chromatin segmentation states, transcription factor binding sites (TFBSs), and histone modification marks using GenomeRunner (http://www.genomerunner.org). P-values were obtained using a Chi-square test and corrected for multiple testing using the Benjamini-Hochberg procedure. We clustered and visualized the ncRNA classes by the strength of their statistical enrichments and depletions.
Conclusions: Searching for statistically significant associations between ncRNA classes and epigenomic elements permits detection of potential functional and/or regulatory relationships among ncRNA classes, and suggests cell type-specific biological roles of ncRNAs.
Figures


References
-
- GEO ftp data archive. ftp://ftp.ncbi.nih.gov/pub/geo/DATA/SOFT/GDS/
-
- ENCODE Project Consortium. The ENCODE (ENCyclopedia Of DNA Elements) Project. Science. 2004;306(5696):636–640. - PubMed
-
- ENCODE Project Consortium. Birney E, Stamatoyannopoulos JA, Dutta A, Guigo R, Gingeras TR, Margulies EH, Weng Z, Snyder M, Dermitzakis ET, Thurman RE. et al.Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447(7146):799–816. doi: 10.1038/nature05874. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

