Dynamics of leading-strand lesion skipping by the replisome
- PMID: 24268579
- PMCID: PMC3877186
- DOI: 10.1016/j.molcel.2013.10.020
Dynamics of leading-strand lesion skipping by the replisome
Abstract
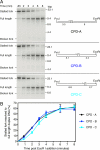
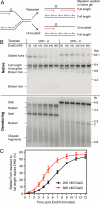
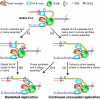
The E. coli replisome stalls transiently when it encounters a lesion in the leading-strand template, skipping over the damage by reinitiating replication at a new primer synthesized downstream by the primase. We report here that template unwinding and lagging-strand synthesis continue downstream of the lesion at a reduced rate after replisome stalling, that one replisome is capable of skipping multiple lesions, and that the rate-limiting steps of replication restart involve the synthesis and activation of the new primer downstream. We also find little support for the concept that polymerase uncoupling, where extensive lagging-strand synthesis proceeds downstream in the absence of leading-strand synthesis, involves physical separation of the leading-strand polymerase from the replisome. Instead, our data indicate that extensive uncoupled replication likely results from a failure of the leading-strand polymerase still associated with the DNA helicase and the lagging-strand polymerase that are proceeding downstream to reinitiate synthesis.
Copyright © 2013 Elsevier Inc. All rights reserved.
Figures


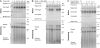

References
-
- Dallmann HG, Kim S, Pritchard AE, Marians KJ, McHenry CS. Characterization of the unique C terminus of the Escherichia coli tau DnaX protein. Monomeric C-tau binds alpha AND DnaB and can partially replace tau in reconstituted replication forks. J. Biol. Chem. 2000;275:15512–15519. - PubMed
-
- Gao D, McHenry CS. tau binds and organizes Escherichia coli replication proteins through distinct domains. Domain IV, located within the unique C terminus of tau, binds the replication fork, helicase, DnaB. J. Biol. Chem. 2001;276:4441–4446. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

