Mechanisms of cohesin-mediated gene regulation and lessons learned from cohesinopathies
- PMID: 24269489
- PMCID: PMC3951616
- DOI: 10.1016/j.bbagrm.2013.11.002
Mechanisms of cohesin-mediated gene regulation and lessons learned from cohesinopathies
Abstract
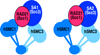
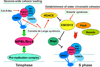
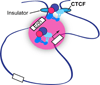
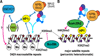
Cohesins are conserved and essential Structural Maintenance of Chromosomes (SMC) protein-containing complexes that physically interact with chromatin and modulate higher-order chromatin organization. Cohesins mediate sister chromatid cohesion and cellular long-distance chromatin interactions affecting genome maintenance and gene expression. Discoveries of mutations in cohesin's subunits and its regulator proteins in human developmental disorders, so-called "cohesinopathies," reveal crucial roles for cohesins in development and cellular growth and differentiation. In this review, we discuss the latest findings concerning cohesin's functions in higher-order chromatin architecture organization and gene regulation and new insight gained from studies of cohesinopathies. This article is part of a Special Issue entitled: Chromatin and epigenetic regulation of animal development.
Keywords: Cohesin; Cohesinopathy; Cornelia de Lange Syndrome; NIPBL; Roberts' Syndrome.
Copyright © 2013 Elsevier B.V. All rights reserved.
Figures
References
-
- Arumugam P, Gruber S, Tanaka K, Haering CH, Mechtler K, Nasmyth K. ATP hydrolysis is required for cohesin’s association with chromosomes. Curr. Biol. 2003;13:1941–1953. - PubMed
-
- Weitzer S, Lehane C, Uhlmann F. A model for ATP hydrolysis-dependent binding of cohesin to DNA. Curr. Biol. 2003;13:1930–1940. - PubMed
-
- Lieb JD, Albrecht MR, Chuang P-T, Meyer BJ. MIX-1: an essential component of the C. elegans mitotic machinery executes X chromosome dosage compensation. Cell. 1998;92:265–277. - PubMed
-
- Hirano T. At the heart of the chromosome: SMC proteins in action. Nat. Rev. Mol. Cell. Biol. 2006;7:311–322. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

