A comparative analysis of trypanosomatid SNARE proteins
- PMID: 24269876
- PMCID: PMC3979113
- DOI: 10.1016/j.parint.2013.11.002
A comparative analysis of trypanosomatid SNARE proteins
Abstract
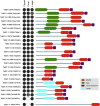
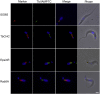
The Kinetoplastida are flagellated protozoa evolutionary distant and divergent from yeast and humans. Kinetoplastida include trypanosomatids, and a number of important pathogens. Trypanosoma brucei, Trypanosoma cruzi and Leishmania spp. inflict significant morbidity and mortality on humans and livestock as the etiological agents of human African trypanosomiasis, Chagas' disease and leishmaniasis respectively. For all of these organisms, intracellular trafficking is vital for maintenance of the host-pathogen interface, modulation/evasion of host immune system responses and nutrient uptake. Soluble N-ethylmaleimide-sensitive factor attachment protein receptors (SNAREs) are critical components of the intracellular trafficking machinery in eukaryotes, mediating membrane fusion and contributing to organelle specificity. We asked how the SNARE complement evolved across the trypanosomatids. An in silico search of the predicted proteomes of T. b. brucei and T. cruzi was used to identify candidate SNARE sequences. Phylogenetic analysis, including comparisons with yeast and human SNAREs, allowed assignment of trypanosomatid SNAREs to the Q or R subclass, as well as identification of several SNAREs orthologous with those of opisthokonts. Only limited variation in number and identity of SNAREs was found, with Leishmania major having 27 and T. brucei 26, suggesting a stable SNARE complement post-speciation. Expression analysis of T. brucei SNAREs revealed significant differential expression between mammalian and insect infective forms, especially within R and Qb-SNARE subclasses, suggesting possible roles in adaptation to different environments. For trypanosome SNAREs with clear orthologs in opisthokonts, the subcellular localization of TbVAMP7C is endosomal while both TbSyn5 and TbSyn16B are at the Golgi complex, which suggests conservation of localization and possibly also function. Despite highly distinct life styles, the complement of trypanosomatid SNAREs is quite stable between the three pathogenic lineages, suggesting establishment in the last common ancestor of trypanosomes and Leishmania. Developmental changes to SNARE mRNA levels between blood steam and procyclic life stages suggest that trypanosomes modulate SNARE functions via expression. Finally, the locations of some conserved SNAREs have been retained across the eukaryotic lineage.
Keywords: Molecular evolution; SNARE; Trypanosoma; Vesicle trafficking.
Copyright © 2013 The Authors. Published by Elsevier Ireland Ltd.. All rights reserved.
Figures





References
-
- Ghedin E., Bringaud F., Peterson J., Myler P., Berriman M., Ivens A. Gene synteny and evolution of genome architecture in trypanosomatids. Mol Biochem Parasitol. 2004;134:183–191. - PubMed
-
- El-Sayed N.M., Myler P.J., Bartholomeu D.C., Nilsson D., Aggarwal G., Tran A.N. The genome sequence of Trypanosoma cruzi, etiologic agent of Chagas disease. Science. 2005;309:409–415. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

