Gene3D: Multi-domain annotations for protein sequence and comparative genome analysis
- PMID: 24270792
- PMCID: PMC3965083
- DOI: 10.1093/nar/gkt1205
Gene3D: Multi-domain annotations for protein sequence and comparative genome analysis
Abstract
Gene3D (http://gene3d.biochem.ucl.ac.uk) is a database of protein domain structure annotations for protein sequences. Domains are predicted using a library of profile HMMs from 2738 CATH superfamilies. Gene3D assigns domain annotations to Ensembl and UniProt sequence sets including >6000 cellular genomes and >20 million unique protein sequences. This represents an increase of 45% in the number of protein sequences since our last publication. Thanks to improvements in the underlying data and pipeline, we see large increases in the domain coverage of sequences. We have expanded this coverage by integrating Pfam and SUPERFAMILY domain annotations, and we now resolve domain overlaps to provide highly comprehensive composite multi-domain architectures. To make these data more accessible for comparative genome analyses, we have developed novel search algorithms for searching genomes to identify related multi-domain architectures. In addition to providing domain family annotations, we have now developed a pipeline for 3D homology modelling of domains in Gene3D. This has been applied to the human genome and will be rolled out to other major organisms over the next year.
Figures
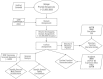
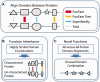

References
-
- Yeats C, Redfern OC, Orengo C. A fast and automated solution for accurately resolving protein domain architectures. Bioinformatics (Oxford, England) 2010;26:745–751. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

