Highly specific and sensitive method for measuring nucleotide excision repair kinetics of ultraviolet photoproducts in human cells
- PMID: 24271390
- PMCID: PMC3936724
- DOI: 10.1093/nar/gkt1179
Highly specific and sensitive method for measuring nucleotide excision repair kinetics of ultraviolet photoproducts in human cells
Abstract
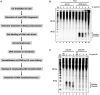
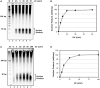
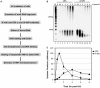
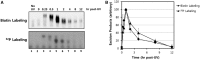
The nucleotide excision repair pathway removes ultraviolet (UV) photoproducts from the human genome in the form of short oligonucleotides ∼ 30 nt in length. Because there are limitations to many of the currently available methods for investigating UV photoproduct repair in vivo, we developed a convenient non-radioisotopic method to directly detect DNA excision repair events in human cells. The approach involves extraction of oligonucleotides from UV-irradiated cells, DNA end-labeling with biotin and streptavidin-mediated chemiluminescent detection of the excised UV photoproduct-containing oligonucleotides that are released from the genome during excision repair. Our novel approach is robust, with essentially no signal in the absence of UV or a functional excision repair system. Furthermore, our non-radioisotopic methodology allows for the sensitive detection of excision products within minutes following UV irradiation and does not require additional enrichment steps such as immunoprecipitation. Finally, this technique allows for quantitative measurements of excision repair in human cells. We suggest that the new techniques presented here will be a useful and powerful approach for studying the mechanism of human nucleotide excision repair in vivo.
Figures


References
-
- Sancar A. DNA excision repair. Annu. Rev. Biochem. 1996;65:43–81. - PubMed
-
- Wood RD. Nucleotide excision repair in mammalian cells. J. Biol. Chem. 1997;272:23465–23468. - PubMed
-
- Reardon JT, Sancar A. Nucleotide excision repair. Prog. Nucleic Acid Res. Mol. Biol. 2005;79:183–235. - PubMed
-
- Svoboda DL, Taylor JS, Hearst JE, Sancar A. DNA repair by eukaryotic nucleotide excision nuclease. Removal of thymine dimer and psoralen monoadduct by HeLa cell-free extract and of thymine dimer by xenopus laevis oocytes. J. Biol. Chem. 1993;268:1931–1936. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

