The impact of equilibrium assumptions on tests of selection
- PMID: 24273554
- PMCID: PMC3822286
- DOI: 10.3389/fgene.2013.00235
The impact of equilibrium assumptions on tests of selection
Abstract
With the increasing availability and quality of whole genome population data, various methodologies of population genetic inference are being utilized in order to identify and quantify recent population-level selective events. Though there has been a great proliferation of such methodology, the type-I and type-II error rates of many proposed statistics have not been well-described. Moreover, the performance of these statistics is often not evaluated for different biologically relevant scenarios (e.g., population size change, population structure), nor for the effect of differing data sizes (i.e., genomic vs. sub-genomic). The absence of the above information makes it difficult to evaluate newly available statistics relative to one another, and thus, difficult to choose the proper toolset for a given empirical analysis. Thus, we here describe and compare the performance of four widely used tests of selection: SweepFinder, SweeD, OmegaPlus, and iHS. In order to consider the above questions, we utilize simulated data spanning a variety of selection coefficients and beneficial mutation rates. We demonstrate that the LD-based OmegaPlus performs best in terms of power to reject the neutral model under both equilibrium and non-equilibrium conditions-an important result regarding the relative effectiveness of linkage disequilibrium relative to site frequency spectrum based statics. The results presented here ought to serve as a useful guide for future empirical studies, and provides a guide for statistical choice depending on the history of the population under consideration. Moreover, the parameter space investigated and the Type-I and Type-II error rates calculated, represent a natural benchmark by which future statistics may be assessed.
Keywords: demography; population genetics; positive selection; simulation; statistical inference.
Figures
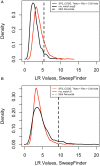
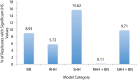
References
-
- Barton N. H. (1998). The effect of hitch-hiking on neutral genealogies. Genet. Res. 72, 123–133 10.1017/S0016672398003462 - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

