Novel orthoreovirus from mink, China, 2011
- PMID: 24274037
- PMCID: PMC3840883
- DOI: 10.3201/eid1912.130043
Novel orthoreovirus from mink, China, 2011
Abstract
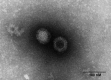
We identified a novel mink orthoreovirus, MRV1HB-A, which seems to be closely related to human strain MRV2tou05, which was isolated from 2 children with acute necrotizing encephalopathy in 2005. Evolution of this virus should be closely monitored so that prevention and control measures can be taken should it become more virulent.
Keywords: China; Orthoreovirus; diarrhea; human; mink; reassortment; reovirus; swine.
Figures

Similar articles
-
Isolation and identification of bat viruses closely related to human, porcine and mink orthoreoviruses.J Gen Virol. 2015 Dec;96(12):3525-3531. doi: 10.1099/jgv.0.000314. J Gen Virol. 2015. PMID: 26475793 Free PMC article.
-
A potentially novel reovirus isolated from swine in northeastern China in 2007.Virus Genes. 2011 Dec;43(3):342-9. doi: 10.1007/s11262-011-0642-4. Epub 2011 Jul 15. Virus Genes. 2011. PMID: 21761235
-
Prevalence and genomic characteristics of a novel reassortment mammalian orthoreovirus type 2 in diarrhea piglets in Sichuan, China.Infect Genet Evol. 2020 Nov;85:104420. doi: 10.1016/j.meegid.2020.104420. Epub 2020 Jun 13. Infect Genet Evol. 2020. PMID: 32544614
-
Novel human reovirus isolated from children with acute necrotizing encephalopathy.Emerg Infect Dis. 2011 Aug;17(8):1436-44. doi: 10.3201/eid1708.101528. Emerg Infect Dis. 2011. PMID: 21801621 Free PMC article.
-
Control of Capsid Transformations during Reovirus Entry.Viruses. 2021 Jan 21;13(2):153. doi: 10.3390/v13020153. Viruses. 2021. PMID: 33494426 Free PMC article. Review.
Cited by
-
Mink, SARS-CoV-2, and the Human-Animal Interface.Front Microbiol. 2021 Apr 1;12:663815. doi: 10.3389/fmicb.2021.663815. eCollection 2021. Front Microbiol. 2021. PMID: 33868218 Free PMC article. Review.
-
Isolation and identification of bat viruses closely related to human, porcine and mink orthoreoviruses.J Gen Virol. 2015 Dec;96(12):3525-3531. doi: 10.1099/jgv.0.000314. J Gen Virol. 2015. PMID: 26475793 Free PMC article.
-
Isolation and characterization of mammalian orthoreovirus type 3 from a fecal sample from a wild boar in Japan.Arch Virol. 2021 Jun;166(6):1671-1680. doi: 10.1007/s00705-021-05053-7. Epub 2021 Apr 11. Arch Virol. 2021. PMID: 33839921
-
Detection of novel orthoreovirus genomes in shrew (Crocidura hirta) and fruit bat (Rousettus aegyptiacus).J Vet Med Sci. 2020 Feb 4;82(2):162-167. doi: 10.1292/jvms.19-0424. Epub 2019 Dec 20. J Vet Med Sci. 2020. PMID: 31866632 Free PMC article.
-
First Specific Detection of Mammalian Orthoreovirus from Goats Using TaqMan Real-Time RT-PCR Technology.Vet Sci. 2024 Mar 22;11(4):141. doi: 10.3390/vetsci11040141. Vet Sci. 2024. PMID: 38668409 Free PMC article.
References
-
- Attoui H, Mertens PPC, Becnel J, Belaganahalli S, Bergoin M, Brussaard CP, et al. Orthoreovirus, Reoviridae. In: Virus taxonomy. Classification and nomenclature of viruses: ninth report of the International Committee on the Taxonomy of Viruses. London: Elsevier Academic Press; 2011. p. 546–54.
-
- Attoui H, Biagini P, Stirling J, Mertens PP, Cantaloube JF, Meyer A, et al. Sequence characterization of Ndelle virus genome segments 1, 5, 7, 8, and 10: evidence for reassignment to the genus Orthoreovirus, family Reoviridae. Biochem Biophys Res Commun. 2001;287:583–8 . 10.1006/bbrc.2001.5612 - DOI - PubMed
Publication types
MeSH terms
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
