Isotopic ratio outlier analysis global metabolomics of Caenorhabditis elegans
- PMID: 24274725
- PMCID: PMC3921967
- DOI: 10.1021/ac4025413
Isotopic ratio outlier analysis global metabolomics of Caenorhabditis elegans
Abstract
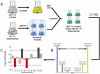
We demonstrate the global metabolic analysis of Caenorhabditis elegans stress responses using a mass-spectrometry-based technique called isotopic ratio outlier analysis (IROA). In an IROA protocol, control and experimental samples are isotopically labeled with 95 and 5% (13)C, and the two sample populations are mixed together for uniform extraction, sample preparation, and LC-MS analysis. This labeling strategy provides several advantages over conventional approaches: (1) compounds arising from biosynthesis are easily distinguished from artifacts, (2) errors from sample extraction and preparation are minimized because the control and experiment are combined into a single sample, (3) measurement of both the molecular weight and the exact number of carbon atoms in each molecule provides extremely accurate molecular formulas, and (4) relative concentrations of all metabolites are easily determined. A heat-shock perturbation was conducted on C. elegans to demonstrate this approach. We identified many compounds that significantly changed upon heat shock, including several from the purine metabolism pathway. The metabolomic response information by IROA may be interpreted in the context of a wealth of genetic and proteomic information available for C. elegans . Furthermore, the IROA protocol can be applied to any organism that can be isotopically labeled, making it a powerful new tool in a global metabolomics pipeline.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

