Soluble expression of proteins correlates with a lack of positively-charged surface
- PMID: 24276756
- PMCID: PMC6506438
- DOI: 10.1038/srep03333
Soluble expression of proteins correlates with a lack of positively-charged surface
Abstract
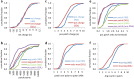
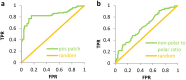
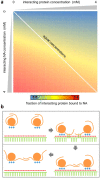
Prediction of protein solubility is gaining importance with the growing use of protein molecules as therapeutics, and ongoing requirements for high level expression. We have investigated protein surface features that correlate with insolubility. Non-polar surface patches associate to some degree with insolubility, but this is far exceeded by the association with positively-charged patches. Negatively-charged patches do not separate insoluble/soluble subsets. The separation of soluble and insoluble subsets by positive charge clustering (area under the curve for a ROC plot is 0.85) has a striking parallel with the separation that delineates nucleic acid-binding proteins, although most of the insoluble dataset are not known to bind nucleic acid. Additionally, these basic patches are enriched for arginine, relative to lysine. The results are discussed in the context of expression systems and downstream processing, contributing to a view of protein solubility in which the molecular interactions of charged groups are far from equivalent.
Conflict of interest statement
The authors declare no competing financial interests.
Figures
References
-
- Wilkinson D. L. & Harrison R. G. Predicting the solubility of recombinant proteins in Escherichia coli. Biotechnology (N Y) 9, 443–448 (1991). - PubMed
-
- Idicula-Thomas S. & Balaji P. V. Correlation between the structural stability and aggregation propensity of proteins. In Silico Biol 7, 225–237 (2007). - PubMed
-
- Smialowski P. et al. Protein solubility: sequence based prediction and experimental verification. Bioinformatics 23, 2536–2542 (2007). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

