The basic leucine zipper transcription factor ABSCISIC ACID RESPONSE ELEMENT-BINDING FACTOR2 is an important transcriptional regulator of abscisic acid-dependent grape berry ripening processes
- PMID: 24276949
- PMCID: PMC3875815
- DOI: 10.1104/pp.113.231977
The basic leucine zipper transcription factor ABSCISIC ACID RESPONSE ELEMENT-BINDING FACTOR2 is an important transcriptional regulator of abscisic acid-dependent grape berry ripening processes
Abstract
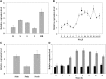
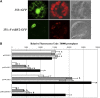
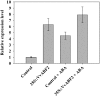
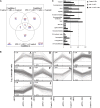
In grape (Vitis vinifera), abscisic acid (ABA) accumulates during fruit ripening and is thought to play a pivotal role in this process, but the molecular basis of this control is poorly understood. This work characterizes ABSCISIC ACID RESPONSE ELEMENT-BINDING FACTOR2 (VvABF2), a grape basic leucine zipper transcription factor belonging to a phylogenetic subgroup previously shown to be involved in ABA and abiotic stress signaling in other plant species. VvABF2 transcripts mainly accumulated in the berry, from the onset of ripening to the harvesting stage, and were up-regulated by ABA. Microarray analysis of transgenic grape cells overexpressing VvABF2 showed that this transcription factor up-regulates and/or modifies existing networks related to ABA responses. In addition, grape cells overexpressing VvABF2 exhibited enhanced responses to ABA treatment compared with control cells. Among the VvABF2-mediated responses highlighted in this study, the synthesis of phenolic compounds and cell wall softening were the most strongly affected. VvABF2 overexpression strongly increased the accumulation of stilbenes that play a role in plant defense and human health (resveratrol and piceid). In addition, the firmness of fruits from tomato (Solanum lycopersicum) plants overexpressing VvABF2 was strongly reduced. These data indicate that VvABF2 is an important transcriptional regulator of ABA-dependent grape berry ripening.
Figures



References
-
- Adams-Phillips L, Barry C, Giovannoni J. (2004) Signal transduction systems regulating fruit ripening. Trends Plant Sci 9: 331–338 - PubMed
-
- Aharoni A, O’Connell AP. (2002) Gene expression analysis of strawberry achene and receptacle maturation using DNA microarrays. J Exp Bot 53: 2073–2087 - PubMed
-
- Amir Hossain M, Lee Y, Cho JI, Ahn CH, Lee SK, Jeon JS, Kang H, Lee CH, An G, Park PB. (2010) The bZIP transcription factor OsABF1 is an ABA responsive element binding factor that enhances abiotic stress signaling in rice. Plant Mol Biol 72: 557–566 - PubMed
-
- Antolín MC, Baigorri H, De Luis I, Aguirrezábal F, Geny L, Broquedis M, Sánchez-Díaz M. (2003) ABA during reproductive development in non-irrigated grapevines (Vitis vinifera L. cv. Tempranillo). Aust J Grape Wine Res 9: 169–176
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

