Disease progression in patients with single, large-scale mitochondrial DNA deletions
- PMID: 24277717
- PMCID: PMC3914470
- DOI: 10.1093/brain/awt321
Disease progression in patients with single, large-scale mitochondrial DNA deletions
Abstract
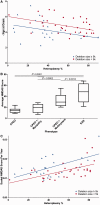
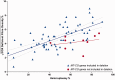
Single, large-scale deletions of mitochondrial DNA are a common cause of mitochondrial disease and cause a broad phenotypic spectrum ranging from mild myopathy to devastating multi-system syndromes such as Kearns-Sayre syndrome. Studies to date have been inconsistent on the value of putative predictors of clinical phenotype and disease progression such as mutation load and the size or location of the deletion. Using a cohort of 87 patients with single, large-scale mitochondrial DNA deletions we demonstrate that a variety of outcome measures such as COX-deficient fibre density, age-at-onset of symptoms and progression of disease burden, as measured by the Newcastle Mitochondrial Disease Adult Scale, are significantly (P < 0.05) correlated with the size of the deletion, the deletion heteroplasmy level in skeletal muscle, and the location of the deletion within the genome. We validate these findings with re-analysis of 256 cases from published data and clarify the previously conflicting information of the value of these predictors, identifying that multiple regression analysis is necessary to understand the effect of these interrelated predictors. Furthermore, we have used mixed modelling techniques to model the progression of disease according to these predictors, allowing a better understanding of the progression over time of this strikingly variable disease. In this way we have developed a new paradigm in clinical mitochondrial disease assessment and management that sidesteps the perennial difficulty of ascribing a discrete clinical phenotype to a broad multi-dimensional and progressive spectrum of disease, establishing a framework to allow better understanding of disease progression.
Keywords: disease progression; mitochondrial DNA deletion; mitochondrial diseases.
Figures



References
-
- Akaike H. A new look at the statistical model identification. IEEE Trans Automat Contr. 1974;AC-19:716–23.
-
- Auré K, Ogier de Baulny H, Laforêt P, Jardel C, Eymard B, Lombès A. Chronic progressive ophthalmoplegia with large-scale mtDNA rearrangement: can we predict progression? Brain. 2007;130:1516–24. - PubMed
-
- Blackwood JK, Whittaker RG, Blakely EL, Alston CL, Turnbull DM, Taylor RW. The investigation and diagnosis of pathogenic mitochondrial DNA mutations in human urothelial cells. Biochem Biophys Res Commun. 2010;393:740–5. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

