The effects of somatic hypermutation on neutralization and binding in the PGT121 family of broadly neutralizing HIV antibodies
- PMID: 24278016
- PMCID: PMC3836729
- DOI: 10.1371/journal.ppat.1003754
The effects of somatic hypermutation on neutralization and binding in the PGT121 family of broadly neutralizing HIV antibodies
Erratum in
- PLoS Pathog. 2013 Dec;9(12). doi:10.1371/annotation/f1f8c791-61e9-45c6-a455-92c6dadf9f06. Kleinstein, Stephen H [corrected to Kleinstein, Steven H]
Abstract
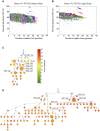
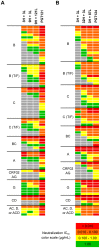
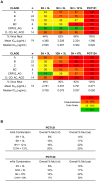
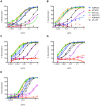
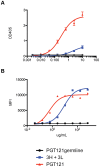
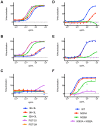
Broadly neutralizing HIV antibodies (bnAbs) are typically highly somatically mutated, raising doubts as to whether they can be elicited by vaccination. We used 454 sequencing and designed a novel phylogenetic method to model lineage evolution of the bnAbs PGT121-134 and found a positive correlation between the level of somatic hypermutation (SHM) and the development of neutralization breadth and potency. Strikingly, putative intermediates were characterized that show approximately half the mutation level of PGT121-134 but were still capable of neutralizing roughly 40-80% of PGT121-134 sensitive viruses in a 74-virus panel at median titers between 15- and 3-fold higher than PGT121-134. Such antibodies with lower levels of SHM may be more amenable to elicitation through vaccination while still providing noteworthy coverage. Binding characterization indicated a preference of inferred intermediates for native Env binding over monomeric gp120, suggesting that the PGT121-134 lineage may have been selected for binding to native Env at some point during maturation. Analysis of glycan-dependent neutralization for inferred intermediates identified additional adjacent glycans that comprise the epitope and suggests changes in glycan dependency or recognition over the course of affinity maturation for this lineage. Finally, patterns of neutralization of inferred bnAb intermediates suggest hypotheses as to how SHM may lead to potent and broad HIV neutralization and provide important clues for immunogen design.
Conflict of interest statement
Authors BBS and EPSJ are employed by 454 Life Sciences - Roche Sequencing Solutions. Authors PYCH and KS are employed by Theraclone-Sciences. FV is employed by, UL and GMC are advisors to, and FV, UL, and GMC own stock in AbVitro, which owns intellectual property in various immune sequencing technologies. The IAVI and Theraclone hold U.S. patent 61/515,528 on the PGT antibodies. All other authors declare no competing interests. This does not alter our adherence to all PLOS Pathogens policies on sharing data and materials.
Figures





References
-
- Kwong PD, Mascola JR (2012) Human antibodies that neutralize HIV-1: identification, structures, and B cell ontogenies. Immunity 37: 412–425 doi:10.1016/j.immuni.2012.08.012 - DOI - PMC - PubMed
-
- Walker LM, Phogat SK, Chan-Hui P-Y, Wagner D, Phung P, et al. (2009) Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science 326: 285–289 doi:10.1126/science.1178746 - DOI - PMC - PubMed
-
- Zhou T, Georgiev I, Wu X, Yang Z-Y, Dai K, et al. (2010) Structural basis for broad and potent neutralization of HIV-1 by antibody VRC01. Science 329: 811–817 doi:10.1126/science.1192819 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- Y1-GM-1104/GM/NIGMS NIH HHS/United States
- 1U19AI090970/AI/NIAID NIH HHS/United States
- T32 AI007606/AI/NIAID NIH HHS/United States
- T32 AI007244/AI/NIAID NIH HHS/United States
- U19 AI090970/AI/NIAID NIH HHS/United States
- 5T32AI007606-10/AI/NIAID NIH HHS/United States
- UM1 AI100663/AI/NIAID NIH HHS/United States
- NIH R01AI033292/AI/NIAID NIH HHS/United States
- R01 AI084817/AI/NIAID NIH HHS/United States
- CAPMC/ CIHR/Canada
- R56 AI084817/AI/NIAID NIH HHS/United States
- AI84817/AI/NIAID NIH HHS/United States
- R01 AI033292/AI/NIAID NIH HHS/United States
- Y1-CO-1020/CO/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources

