HDAC4 does not act as a protein deacetylase in the postnatal murine brain in vivo
- PMID: 24278330
- PMCID: PMC3838388
- DOI: 10.1371/journal.pone.0080849
HDAC4 does not act as a protein deacetylase in the postnatal murine brain in vivo
Abstract
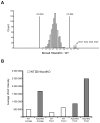
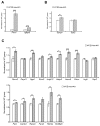
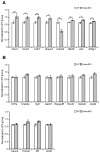
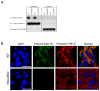
Reversible protein acetylation provides a central mechanism for controlling gene expression and cellular signaling events. It is governed by the antagonistic commitment of two enzymes families: the histone acetyltransferases (HATs) and the histone deacetylases (HDACs). HDAC4, like its class IIa counterparts, is a potent transcriptional repressor through interactions with tissue specific transcription factors via its N-terminal domain. Whilst the lysine deacetylase activity of the class IIa HDACs is much less potent than that of the class I enzymes, HDAC4 has been reported to influence protein deacetylation through its interaction with HDAC3. To investigate the influence of HDAC4 on protein acetylation we employed the immunoaffinity-based AcetylScan proteomic method. We identified many proteins known to be modified by acetylation, but found that the absence of HDAC4 had no effect on the acetylation profile of the murine neonate brain. This is consistent with the biochemical data suggesting that HDAC4 may not function as a lysine deacetylase, but these in vivo data do not support the previous report showing that the enzymatic activity of HDAC3 might be modified by its interaction with HDAC4. To complement this work, we used Affymetrix arrays to investigate the effect of HDAC4 knock-out on the transcriptional profile of the postnatal murine brain. There was no effect on global transcription, consistent with the absence of a differential histone acetylation profile. Validation of the array data by Taq-man qPCR indicated that only protamine 1 and Igfbp6 mRNA levels were increased by more than one-fold and only Calml4 was decreased. The lack of a major effect on the transcriptional profile is consistent with the cytoplasmic location of HDAC4 in the P3 murine brain.
Conflict of interest statement
Figures
References
-
- McKinsey TA, Zhang CL, Olson EN (2000) Activation of the myocyte enhancer factor-2 transcription factor by calcium/calmodulin-dependent protein kinase-stimulated binding of 14-3-3 to histone deacetylase 5. Proc Natl Acad Sci U S A 97: 14400-14405. doi:10.1073/pnas.260501497. PubMed: 11114197. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

