Osmostress-induced cell volume loss delays yeast Hog1 signaling by limiting diffusion processes and by Hog1-specific effects
- PMID: 24278344
- PMCID: PMC3835318
- DOI: 10.1371/journal.pone.0080901
Osmostress-induced cell volume loss delays yeast Hog1 signaling by limiting diffusion processes and by Hog1-specific effects
Abstract
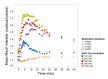
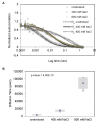
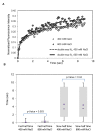
Signal transmission progresses via a series of transient protein-protein interactions and protein movements, which require diffusion within a cell packed with different molecules. Yeast Hog1, the effector protein kinase of the High Osmolarity Glycerol pathway, translocates transiently from the cytosol to the nucleus during adaptation to high external osmolarity. We followed the dynamics of osmostress-induced cell volume loss and Hog1 nuclear accumulation upon exposure of cells to different NaCl concentrations. While Hog1 nuclear accumulation peaked within five minutes following mild osmotic shock it was delayed up to six-fold under severe stress. The timing of Hog1 nuclear accumulation correlated with the degree of cell volume loss and the cells capacity to recover. Also the nuclear translocation of Msn2, the transcription factor of the general stress response pathway, is delayed upon severe osmotic stress suggesting a general phenomenon. We show by direct measurements that the general diffusion rate of Hog1 in the cytoplasm as well as its rate of nuclear transport are dramatically reduced following severe volume reduction. However, neither Hog1 phosphorylation nor Msn2 nuclear translocation were as much delayed as Hog1 nuclear translocation. Our data provide direct evidence that signaling slows down during cell volume compression, probably as a consequence of molecular crowding. Hence one purpose of osmotic adaptation is to restore optimal diffusion rates for biochemical and cell biological processes. In addition, there may be mechanisms slowing down especially Hog1 nuclear translocation under severe stress in order to prioritize Hog1 cytosolic targets.
Conflict of interest statement
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

