Investigations on the Tobacco Necrosis Virus D p60 replicase protein
- PMID: 24278346
- PMCID: PMC3836746
- DOI: 10.1371/journal.pone.0080912
Investigations on the Tobacco Necrosis Virus D p60 replicase protein
Abstract
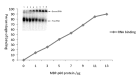
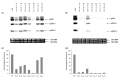
Tobacco Necrosis Virus D (TNV-D), in the genus Betanecrovirus (family Tombusviridae), possesses a single-stranded, positive-sense RNA genome containing six open reading frames (ORFs). Two 5'-proximal ORFs (1 and 2) encode overlapping polypeptides of 22 and 82 kDa (p22 and p82, respectively) which are both required for replication. The p22 auxiliary protein contains no replication motifs but the C-terminal region, downstream of a leaky stop codon, encodes a 60 kDa polypeptide (p60) which contains conserved RNA-dependent RNA polymerase (RdRP) motifs. Here we have expressed and purified recombinant p60 and show that in vitro it binds and efficiently synthesises both TNV-D RNA and Satellite tobacco necrosis virus C RNA. Alanine scanning mutagenesis of conserved amino acids in characteristic motifs in p60 revealed that some mutations significantly reduced RNA synthesis but mutating the second asparagine residue in the conserved GDD box was lethal. The effects of mutating identical amino acids in p60 on virus replication in vivo were examined in Nicotiana benthamiana plants following infection with RNA transcribed from wild type (wt) and mutant constructs. In inoculated leaves the behaviour of the mutants paralleled the in vitro data but systemic infection was precluded in all but one mutant which had reverted to wt. This study is the first to demonstrate the nucleic acid-binding and synthetic capabilities of a betanecrovirus polymerase.
Conflict of interest statement
Figures



References
-
- Rochon D, Lommel SA, Martelli GP, Rubino L, Russo M (2012) Tombusviridae. In King, A.M.Q.; Adams M.J., Carstens E.B., Lefkowitz E.J., (Eds.).Virus Taxonomy, Ninth Report of the International Committee on Taxonomy of Viruses, Elsevier / Academic Press, London: , pp 1111–1138
-
- Offei SK, Coutts RHA (1996) Location of the 5’-termini of the tobacco necrosis virus strain D subgenomic RNAs. J Phytopathol 144: 13-17. doi:10.1111/j.1439-0434.1996.tb01481.x. - DOI
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

