Recognizing the enemy within: licensing RNA-guided genome defense
- PMID: 24280023
- PMCID: PMC3902128
- DOI: 10.1016/j.tibs.2013.10.003
Recognizing the enemy within: licensing RNA-guided genome defense
Abstract
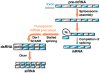
How do cells distinguish normal genes from transposons? Although much has been learned about RNAi-related RNA silencing pathways responsible for genome defense, this fundamental question remains. The literature points to several classes of mechanisms. In some cases, double-stranded RNA (dsRNA) structures produced by transposon inverted repeats or antisense integration trigger endogenous small interfering RNA (siRNA) biogenesis. In other instances, DNA features associated with transposons--such as their unusual copy number, chromosomal arrangement, and/or chromatin environment--license RNA silencing. Finally, recent studies have identified improper transcript processing events, such as stalled pre-mRNA splicing, as signals for siRNA production. Thus, the suboptimal gene expression properties of selfish elements can enable their identification by RNA silencing pathways.
Keywords: PIWI-interacting RNA; RNAi; genome defense; small RNA; small interfering RNA; transposon.
Copyright © 2013 Elsevier Ltd. All rights reserved.
Figures


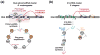
Similar articles
-
Transposon silencing in the Caenorhabditis elegans germ line by natural RNAi.Nature. 2003 Nov 20;426(6964):310-4. doi: 10.1038/nature02107. Nature. 2003. PMID: 14628056
-
Fighting an old war with a new weapon--silencing transposons by Piwi-interacting RNA.IUBMB Life. 2013 Sep;65(9):739-47. doi: 10.1002/iub.1192. Epub 2013 Jul 29. IUBMB Life. 2013. PMID: 23893818 Review.
-
A double-stranded-RNA response program important for RNA interference efficiency.Mol Cell Biol. 2007 Jun;27(11):3995-4005. doi: 10.1128/MCB.00186-07. Epub 2007 Mar 19. Mol Cell Biol. 2007. PMID: 17371837 Free PMC article.
-
Inhibition of RNA interference and modulation of transposable element expression by cell death in Drosophila.Genetics. 2011 Aug;188(4):823-34. doi: 10.1534/genetics.111.128470. Epub 2011 May 19. Genetics. 2011. PMID: 21596898 Free PMC article.
-
Conserved themes in small-RNA-mediated transposon control.Trends Cell Biol. 2008 Mar;18(3):136-48. doi: 10.1016/j.tcb.2008.01.004. Epub 2008 Feb 20. Trends Cell Biol. 2008. PMID: 18282709 Free PMC article. Review.
Cited by
-
Epigenetic control of mobile DNA as an interface between experience and genome change.Front Genet. 2014 Apr 25;5:87. doi: 10.3389/fgene.2014.00087. eCollection 2014. Front Genet. 2014. PMID: 24795749 Free PMC article. Review.
-
Nuclear Noncoding RNAs and Genome Stability.Mol Cell. 2016 Jul 7;63(1):7-20. doi: 10.1016/j.molcel.2016.06.011. Mol Cell. 2016. PMID: 27392145 Free PMC article. Review.
-
A Small RNA-Based Immune System Defends Germ Cells against Mobile Genetic Elements.Stem Cells Int. 2016;2016:7595791. doi: 10.1155/2016/7595791. Epub 2015 Nov 22. Stem Cells Int. 2016. PMID: 26681955 Free PMC article. Review.
-
Putative RNA-directed adaptive mutations in cancer evolution.Transcription. 2016 Oct 19;7(5):164-187. doi: 10.1080/21541264.2016.1221491. Transcription. 2016. PMID: 27715501 Free PMC article.
-
Spotting the enemy within: Targeted silencing of foreign DNA in mammalian genomes by the Krüppel-associated box zinc finger protein family.Mob DNA. 2015 Oct 2;6:17. doi: 10.1186/s13100-015-0050-8. eCollection 2015. Mob DNA. 2015. PMID: 26435754 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

