Narrowing down the targets for yield improvement in rice under normal and abiotic stress conditions via expression profiling of yield-related genes
- PMID: 24280046
- PMCID: PMC4883727
- DOI: 10.1186/1939-8433-5-37
Narrowing down the targets for yield improvement in rice under normal and abiotic stress conditions via expression profiling of yield-related genes
Abstract
Background: Crop improvement targeting high yield and tolerance to environmental stresses has become the need of the hour. Yield improvement via breeding or gene pyramiding aiming comprehensive incorporation of the agronomically favored traits requires an in-depth understanding of the molecular basis of these traits. The present study describes expression profiling of yield-related genes in rice with respect to different developmental stages and various abiotic stress conditions.
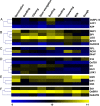
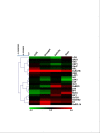
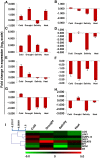
Results: Our analysis indicates developmental regulation of the yield-related genes pertaining to the genetic reprogramming involved at the corresponding developmental stage. The gene expression data can be utilized to specifically select particular genes which can potentially function synergistically for enhancing the yield while maintaining the source-sink balance. Furthermore, to gain some insights into the molecular basis of yield penalty during various abiotic stresses, the expression of selected yield-related genes has also been analyzed by qRT-PCR under such stress conditions. Our analysis clearly showed a tight transcriptional regulation of a few of these yield-related genes by abiotic stresses. The stress-responsive expression patterns of these genes could explain some of the most important stress-related physiological manifestations such as reduced tillering, smaller panicles and early completion of the life cycle owing to reduced duration of vegetative and reproductive phases.
Conclusions: Development of high yielding rice varieties which maintain their yield even under stress conditions may be achieved by simultaneous genetic manipulation of certain combination of genes such as LRK1 and LOG, based on their function and expression profile obtained in the present study. Our study would aid in investigating in future, whether over-expressing or knocking down such yield-related genes can improve the grain yield potential in rice.
Figures

References
-
- Hiei Y, Komari T. Improved protocols for transformation of indica rice mediated by Agrobacterium tumefaciens. Plant Cell, Tissue and Organ cult. 2006;85:271–554. doi: 10.1007/s11240-005-9069-8. - DOI
LinkOut - more resources
Full Text Sources

