Insights into genomics of salt stress response in rice
- PMID: 24280112
- PMCID: PMC4883734
- DOI: 10.1186/1939-8433-6-27
Insights into genomics of salt stress response in rice
Abstract
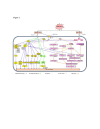
Plants, as sessile organisms experience various abiotic stresses, which pose serious threat to crop production. Plants adapt to environmental stress by modulating their growth and development along with the various physiological and biochemical changes. This phenotypic plasticity is driven by the activation of specific genes encoding signal transduction, transcriptional regulation, ion transporters and metabolic pathways. Rice is an important staple food crop of nearly half of the world population and is well known to be a salt sensitive crop. The completion and enhanced annotations of rice genome sequence has provided the opportunity to study functional genomics of rice. Functional genomics aids in understanding the molecular and physiological basis to improve the salinity tolerance for sustainable rice production. Salt tolerant transgenic rice plants have been produced by incorporating various genes into rice. In this review we present the findings and investigations in the field of rice functional genomics that includes supporting genes and networks (ABA dependent and independent), osmoprotectants (proline, glycine betaine, trehalose, myo-inositol, and fructans), signaling molecules (Ca2+, abscisic acid, jasmonic acid, brassinosteroids) and transporters, regulating salt stress response in rice.
Figures


Similar articles
-
Bacillus amyloliquefaciens Confers Tolerance to Various Abiotic Stresses and Modulates Plant Response to Phytohormones through Osmoprotection and Gene Expression Regulation in Rice.Front Plant Sci. 2017 Aug 29;8:1510. doi: 10.3389/fpls.2017.01510. eCollection 2017. Front Plant Sci. 2017. PMID: 28900441 Free PMC article.
-
Microarray Analysis of Rice d1 (RGA1) Mutant Reveals the Potential Role of G-Protein Alpha Subunit in Regulating Multiple Abiotic Stresses Such as Drought, Salinity, Heat, and Cold.Front Plant Sci. 2016 Jan 28;7:11. doi: 10.3389/fpls.2016.00011. eCollection 2016. Front Plant Sci. 2016. PMID: 26858735 Free PMC article.
-
Enhancement of Plant Productivity in the Post-Genomics Era.Curr Genomics. 2016 Aug;17(4):295-6. doi: 10.2174/138920291704160607182507. Curr Genomics. 2016. PMID: 27499678 Free PMC article.
-
Brassinosteroids: A Promising Option in Deciphering Remedial Strategies for Abiotic Stress Tolerance in Rice.Front Plant Sci. 2017 Dec 20;8:2151. doi: 10.3389/fpls.2017.02151. eCollection 2017. Front Plant Sci. 2017. PMID: 29326745 Free PMC article. Review.
-
A Review of Integrative Omic Approaches for Understanding Rice Salt Response Mechanisms.Plants (Basel). 2022 May 27;11(11):1430. doi: 10.3390/plants11111430. Plants (Basel). 2022. PMID: 35684203 Free PMC article. Review.
Cited by
-
Chloroplast Localized FIBRILLIN11 Is Involved in the Osmotic Stress Response during Arabidopsis Seed Germination.Biology (Basel). 2021 Apr 25;10(5):368. doi: 10.3390/biology10050368. Biology (Basel). 2021. PMID: 33922967 Free PMC article.
-
Azospirillum brasilense improves rice growth under salt stress by regulating the expression of key genes involved in salt stress response, abscisic acid signaling, and nutrient transport, among others.Front Agron. 2023;5:1216503. doi: 10.3389/fagro.2023.1216503. Epub 2023 Oct 4. Front Agron. 2023. PMID: 38223701 Free PMC article.
-
Overexpression of Rice Expansin7 (Osexpa7) Confers Enhanced Tolerance to Salt Stress in Rice.Int J Mol Sci. 2020 Jan 10;21(2):454. doi: 10.3390/ijms21020454. Int J Mol Sci. 2020. PMID: 31936829 Free PMC article.
-
Comparative transcriptome analysis reveals K+ transporter gene contributing to salt tolerance in eggplant.BMC Plant Biol. 2019 Feb 11;19(1):67. doi: 10.1186/s12870-019-1663-8. BMC Plant Biol. 2019. PMID: 30744551 Free PMC article.
-
Advances in understanding salt tolerance in rice.Theor Appl Genet. 2019 Apr;132(4):851-870. doi: 10.1007/s00122-019-03301-8. Epub 2019 Feb 13. Theor Appl Genet. 2019. PMID: 30759266 Review.
References
-
- Agrawal GK, Rakwa R, Iwahashi H. Isolation of novel rice (Oryza sativa L.) multiple stress responsive MAP kinase gene, OsMSRMK2, whose mRNA accumulates rapidly in response to environmental cues. Biochem Biophys Res Commun. 2002;294:1009–1016. - PubMed
-
- Agrawal GK, Agrawal SK, Shibato J, Iwahashi H, Rakwal R. Novel rice MAP kinases OsMSRMK3 and OsWJUMK1 involved in encountering diverse environmental stresses and developmental regulation. Biochem Biophys Res Commun. 2003;300:775–783. - PubMed
-
- Asano T, Hakata M, Nakamura H, Aoki N, Komatsu S, Ichikawa H, Hirochika H, Ohsugi R. Functional characterisation of OsCPK21, a calcium-dependent protein kinase that confers salt tolerance in rice. Plant Mol Biol. 2011;75:179–191. - PubMed
-
- Asano T, Hayashi N, Kobayashi M, Aoki N, Miyao A, Mitsuhara I, Ichikawa H, Komatsu S, Hirochika H, Kikuchi S, Ohsugi R. A rice calcium-dependent protein kinase OsCPK12 oppositely modulates salt-stress tolerance and blast disease resistance. Plant J. 2012;69:26–36. - PubMed
-
- Ashraf M, Foolad MR. Roles of glycine betaine and proline in improving plant abiotic stress resistance. Environ Exp Bot. 2007;59:206–216.
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

