Genome-wide identification and analysis of miRNA-related single nucleotide polymorphisms (SNPs) in rice
- PMID: 24280131
- PMCID: PMC4883715
- DOI: 10.1186/1939-8433-6-10
Genome-wide identification and analysis of miRNA-related single nucleotide polymorphisms (SNPs) in rice
Abstract
Background: MiRNAs are key regulators in the miRNA-mediated regulatory networks. Single nucleotide polymorphisms (SNPs) that occur at miRNA-related regions may cause serious phenotype changes. To gain new insights into the evolution of miRNAs after SNP variation, we performed a genome-wide scan of miRNA-related SNPs, and analyzed their effects on the stability of miRNAs structure and the alteration of target spectrum in rice.
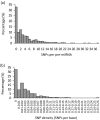
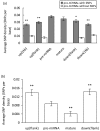
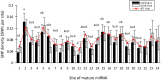
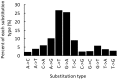
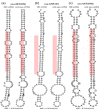
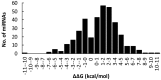
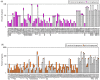
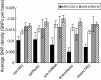
Results: We find that the SNP density in pre-miRNAs is significantly higher than that in the flanking regions, owing to the rapid evolution of a large number of species-specific miRNAs in rice. In contrast, it is obvious that deeply conserved miRNAs are under strong purifying selection during evolution. In most cases, the SNPs in stem regions may result in the miRNA hairpin structures changing from stable to unstable status; And SNPs in mature miRNAs have great potential to have either newly created or disrupted the miRNA-target interactions. However, the total number of gained targets is over 2.5 times greater than that are lost after mutation. Notably, 12 putative domestication-related miRNAs have been identified, where the SNP density is significantly lower.
Conclusions: The present study provides the first outline of SNP variations occurred in rice pre-miRNAs at the whole genome-wide level. These analyses may deepen our understanding on the effects of SNPs on the evolution of miRNAs in the rice genome.
Figures
Similar articles
-
Genome-Wide Analysis of MicroRNA-related Single Nucleotide Polymorphisms (SNPs) in Mouse Genome.Sci Rep. 2020 Apr 1;10(1):5789. doi: 10.1038/s41598-020-62588-6. Sci Rep. 2020. PMID: 32238847 Free PMC article.
-
Genome-wide identification of SNPs in microRNA genes and the SNP effects on microRNA target binding and biogenesis.Hum Mutat. 2012 Jan;33(1):254-63. doi: 10.1002/humu.21641. Epub 2011 Nov 23. Hum Mutat. 2012. PMID: 22045659
-
Role of SNPs in the Biogenesis of Mature miRNAs.Biomed Res Int. 2021 Jun 17;2021:2403418. doi: 10.1155/2021/2403418. eCollection 2021. Biomed Res Int. 2021. PMID: 34239922 Free PMC article.
-
[Single nucleotide polymorphism (SNP) and its application in rice].Yi Chuan. 2006 Jun;28(6):737-44. Yi Chuan. 2006. PMID: 16818440 Review. Chinese.
-
miRNAs, single nucleotide polymorphisms (SNPs) and age-related macular degeneration (AMD).Clin Chem Lab Med. 2017 May 1;55(5):763-775. doi: 10.1515/cclm-2016-0898. Clin Chem Lab Med. 2017. PMID: 28343170 Review.
Cited by
-
Single nucleotide polymorphisms affect miRNA target prediction in bovine.PLoS One. 2021 Apr 21;16(4):e0249406. doi: 10.1371/journal.pone.0249406. eCollection 2021. PLoS One. 2021. PMID: 33882076 Free PMC article.
-
Expression Variations of miRNAs and mRNAs in Rice (Oryza sativa).Genome Biol Evol. 2016 Dec 31;8(11):3529-3544. doi: 10.1093/gbe/evw252. Genome Biol Evol. 2016. PMID: 27797952 Free PMC article.
-
Genome-wide identification and evolutionary analysis of positively selected miRNA genes in domesticated rice.Mol Genet Genomics. 2015 Apr;290(2):593-602. doi: 10.1007/s00438-014-0943-0. Epub 2014 Nov 2. Mol Genet Genomics. 2015. PMID: 25362560
-
Comparative analysis of the Dicer-like gene family reveals loss of miR162 target site in SmDCL1 from Salvia miltiorrhiza.Sci Rep. 2015 May 13;5:9891. doi: 10.1038/srep09891. Sci Rep. 2015. PMID: 25970825 Free PMC article.
-
Development of miRNA-SSR and target-SSR markers from yield-associate genes and their applicability in the assessment of genetic diversity and association mapping in rice (Oryza sativa L.).Mol Breed. 2024 Apr 15;44(4):30. doi: 10.1007/s11032-024-01462-z. eCollection 2024 Apr. Mol Breed. 2024. PMID: 38634111 Free PMC article.
References
-
- Atwell S, Huang YS, Vilhjálmsson BJ, Willems G, Horton M, Li Y, Meng D, Platt A, Tarone AW, Hu TT, Jiang R, Muliyati NW, Zhang X, Amer MA, Baxter I, Brachi B, Chory J, Dean C, Debieu M, De Meaux J, Ecker JR, Faure N, Kniskern JM, Jones JD, Michael T, Nemri A, Roux F, Salt DE, Tang C, Todesco M, Traw MB, Weigel D, Marjoram P, Borevitz JO, Bergelson J, Nordborg M. Genome-wide association study of 107 phenotypes in Arabidopsis thaliana inbred lines. Nature. 2010;465:627–631. doi: 10.1038/nature08800. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

