Identification of genes regulating growth and fatness traits in pig through hypothalamic transcriptome analysis
- PMID: 24280257
- PMCID: PMC3949103
- DOI: 10.1152/physiolgenomics.00151.2013
Identification of genes regulating growth and fatness traits in pig through hypothalamic transcriptome analysis
Abstract
Previous studies on Iberian × Landrace (IBMAP) pig intercrosses have enabled the identification of several quantitative trait locus (QTL) regions related to growth and fatness traits; however, the genetic variation underlying those QTLs are still unknown. These traits are not only relevant because of their impact on economically important production traits, but also because pig constitutes a widely studied animal model for human obesity and obesity-related diseases. The hypothalamus is the main gland regulating growth, food intake, and fat accumulation. Therefore, the aim of this work was to identify genes and/or gene transcripts involved in the determination of growth and fatness in pig by a comparison of the whole hypothalamic transcriptome (RNA-Seq) in two groups of phenotypically divergent IBMAP pigs. Around 16,000 of the ∼25.010 annotated genes were expressed in these hypothalamic samples, with most of them showing intermediate expression levels. Functional analyses supported the key role of the hypothalamus in the regulation of growth, fat accumulation, and energy expenditure. Moreover, 58,927 potentially new isoforms were detected. More than 250 differentially expressed genes and novel transcript isoforms were identified between the two groups of pigs. Twenty-one DE genes/transcripts that colocalized in previously identified QTL regions and/or whose biological functions are related to the traits of interest were explored in more detail. Additionally, the transcription factors potentially regulating these genes and the subjacent networks and pathways were also analyzed. This study allows us to propose strong candidate genes for growth and fatness based on expression patterns, genomic location, and network interactions.
Keywords: RNA-Seq; fatness; growth; hypothalamus; porcine.
Figures
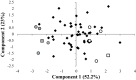
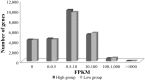


Similar articles
-
Comparative adipose transcriptome analysis digs out genes related to fat deposition in two pig breeds.Sci Rep. 2019 Sep 9;9(1):12925. doi: 10.1038/s41598-019-49548-5. Sci Rep. 2019. PMID: 31501489 Free PMC article.
-
Genetics of fat tissue accumulation in pigs: a comparative approach.J Appl Genet. 2010;51(2):153-68. doi: 10.1007/BF03195724. J Appl Genet. 2010. PMID: 20453303
-
Analysis of porcine adipose tissue transcriptome reveals differences in de novo fatty acid synthesis in pigs with divergent muscle fatty acid composition.BMC Genomics. 2013 Dec 1;14:843. doi: 10.1186/1471-2164-14-843. BMC Genomics. 2013. PMID: 24289474 Free PMC article.
-
Genome-wide association analyses reveal significant loci and strong candidate genes for growth and fatness traits in two pig populations.Genet Sel Evol. 2015 Mar 14;47(1):17. doi: 10.1186/s12711-015-0089-5. Genet Sel Evol. 2015. PMID: 25885760 Free PMC article.
-
Deciphering the regulation of porcine genes influencing growth, fatness and yield-related traits through genetical genomics.Mamm Genome. 2017 Apr;28(3-4):130-142. doi: 10.1007/s00335-016-9674-3. Epub 2016 Dec 10. Mamm Genome. 2017. PMID: 27942838
Cited by
-
Identification of Candidate Genes and Regulatory Factors Underlying Intramuscular Fat Content Through Longissimus Dorsi Transcriptome Analyses in Heavy Iberian Pigs.Front Genet. 2018 Dec 4;9:608. doi: 10.3389/fgene.2018.00608. eCollection 2018. Front Genet. 2018. PMID: 30564273 Free PMC article.
-
Extensive expression differences along porcine small intestine evidenced by transcriptome sequencing.PLoS One. 2014 Feb 12;9(2):e88515. doi: 10.1371/journal.pone.0088515. eCollection 2014. PLoS One. 2014. PMID: 24533095 Free PMC article.
-
A phylogenomic analysis of the role and timing of molecular adaptation in the aquatic transition of cetartiodactyl mammals.R Soc Open Sci. 2015 Sep 30;2(9):150156. doi: 10.1098/rsos.150156. eCollection 2015 Sep. R Soc Open Sci. 2015. PMID: 26473040 Free PMC article.
-
Transcriptome Analysis in Domesticated Species: Challenges and Strategies.Bioinform Biol Insights. 2016 Feb 16;9(Suppl 4):21-31. doi: 10.4137/BBI.S29334. eCollection 2015. Bioinform Biol Insights. 2016. PMID: 26917953 Free PMC article. Review.
-
Slc12a2 loss in insulin-secreting β-cells links development of overweight and metabolic dysregulation to impaired satiation control of feeding.Am J Physiol Endocrinol Metab. 2023 Nov 1;325(5):E581-E594. doi: 10.1152/ajpendo.00197.2023. Epub 2023 Oct 11. Am J Physiol Endocrinol Metab. 2023. PMID: 37819196 Free PMC article.
References
-
- Barb CR, Hausman GJ, Rekaya R, Lents CA, Lkhagvadorj S, Qu L, Cai W, Couture OP, Anderson LL, Dekkers JC, Tuggle CK. Gene expression in hypothalamus, liver and adipose tissues and food intake response to melanocortin-4 receptor (MC4R) agonist in pigs expressing MC4R mutations. Physiol Genomics 41: 254–268, 2010. - PubMed
-
- Cellini E, Castellini G, Ricca V, Bagnoli S, Tedde A, Rotella CM, Faravelli C, Sorbi S, Nacmias B. Glucocorticoid receptor gene polymorphisms in Italian patients with eating disorders and obesity. Psychiatr Genet 20: 282–288, 2010. - PubMed
-
- Currie RA, Eckel RH. Characterization of a high affinity octamer transcription factor binding site in the human lipoprotein lipase promoter. Arch Biochem Biophys 298: 630–639, 1992. - PubMed
Publication types
MeSH terms
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

