Diversity and genetics of nitrogen-induced susceptibility to the blast fungus in rice and wheat
- PMID: 24280346
- PMCID: PMC4883689
- DOI: 10.1186/1939-8433-6-32
Diversity and genetics of nitrogen-induced susceptibility to the blast fungus in rice and wheat
Abstract
Background: Nitrogen often increases disease susceptibility, a phenomenon that can be observed under controlled conditions and called NIS, for Nitrogen-Induced Susceptibility. NIS has long been reported in the case of rice blast disease caused by the fungus Magnaporthe oryzae. We used an experimental system that does not strongly affect plant development to address the question of NIS polymorphism across rice diversity and further explored this phenomenon in wheat. We tested the two major types of resistance, namely quantitative/partial resistance and resistance driven by known resistance genes. Indeed there are conflicting reports on the effects of NIS on the first one and none on the last one. Finally, the genetics of NIS is not well documented and only few loci have been identified that may control this phenomenon.
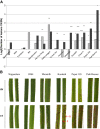
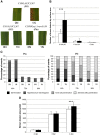
Results: Our data indicate that NIS is a general phenomenon affecting resistance to blast fungus in these two cereals. We show that the capacity of rice to display NIS is highly polymorphic and does not correlate with difference related to indica/japonica sub-groups. We also tested the robustness of three different major resistance genes under high nitrogen. Nitrogen partially breaks down resistance triggered by the Pi1 gene. Cytological examination indicates that penetration rate is not affected by high nitrogen whereas growth of the fungus is increased inside the plant. Using the CSSL mapping population between Nipponbare and Kasalath, we identified a Kasalath locus on chromosome 1, called NIS1, which dominantly increases susceptibility under high nitrogen. We discuss the possible relationships between Nitrogen Use Efficiency (NUE), disease resistance regulation and NIS.
Conclusions: This work provides evidences that robust forms of partial resistance exist across diversity and can be easily identified with our protocol. This work also suggests that under certain environmental circumstances, complete resistance may breakdown, irrelevantly of the capacity of the fungus to mutate. These aspects should be considered while breeding for robust forms of resistance to blast disease.
Figures



References
-
- Ballini E, Morel J-B, Droc G, Price A, Courtois B, Notteghem J-L, Tharreau D. A genome-wide meta-analysis of rice blast resistance genes and quantitative trait loci provides new insights into partial and complete resistance. Mol Plant Microb Interact. 2008;21:859–868. doi: 10.1094/MPMI-21-7-0859. - DOI - PubMed
-
- Berruyer R, Adreit H, Milazzo J, Gaillard S, Berger A, Dioh W, Lebrun MH, Tharreau D. Identification and fine mapping of Pi33, the rice resistance gene corresponding to the Magnaporthe grisea avirulence gene ACE1. Theor Appl Genet. 2003;107(612):1139–1147. doi: 10.1007/s00122-003-1349-2. - DOI - PubMed
-
- Bonman JM. Durable resistance to rice blast disease - environmental influences. Euphytica. 1992;63:115–123. doi: 10.1007/BF00023917. - DOI
-
- Cesari S, Thilliez G, Ribot C, Chalvon V, Michel C, Jauneau A, Rivas S, Alaux L, Kanzaki H, Okuyama Y, Morel J-B, Fournier E, Tharreau D, Terauchi R, Kroj T. The rice resistance protein pair RGA4/RGA5 recognizes the Magnaporthe oryzae effectors AVR-Pia and AVR1-CO39 by direct binding. Plant Cell Online. 2013;25(4):1463–1481. doi: 10.1105/tpc.112.107201. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

