Systems-level analysis of nitrogen starvation-induced modifications of carbon metabolism in a Chlamydomonas reinhardtii starchless mutant
- PMID: 24280389
- PMCID: PMC3875720
- DOI: 10.1105/tpc.113.117580
Systems-level analysis of nitrogen starvation-induced modifications of carbon metabolism in a Chlamydomonas reinhardtii starchless mutant
Abstract
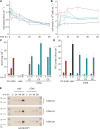
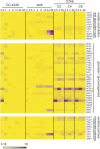
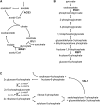
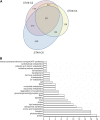
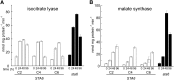
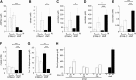
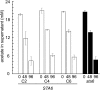
To understand the molecular basis underlying increased triacylglycerol (TAG) accumulation in starchless (sta) Chlamydomonas reinhardtii mutants, we undertook comparative time-course transcriptomics of strains CC-4348 (sta6 mutant), CC-4349, a cell wall-deficient (cw) strain purported to represent the parental STA6 strain, and three independent STA6 strains generated by complementation of sta6 (CC-4565/STA6-C2, CC-4566/STA6-C4, and CC-4567/STA6-C6) in the context of N deprivation. Despite N starvation-induced dramatic remodeling of the transcriptome, there were relatively few differences (5 × 10(2)) observed between sta6 and STA6, the most dramatic of which were increased abundance of transcripts encoding key regulated or rate-limiting steps in central carbon metabolism, specifically isocitrate lyase, malate synthase, transaldolase, fructose bisphosphatase and phosphoenolpyruvate carboxykinase (encoded by ICL1, MAS1, TAL1, FBP1, and PCK1 respectively), suggestive of increased carbon movement toward hexose-phosphate in sta6 by upregulation of the glyoxylate pathway and gluconeogenesis. Enzyme assays validated the increase in isocitrate lyase and malate synthase activities. Targeted metabolite analysis indicated increased succinate, malate, and Glc-6-P and decreased Fru-1,6-bisphosphate, illustrating the effect of these changes. Comparisons of independent data sets in multiple strains allowed the delineation of a sequence of events in the global N starvation response in C. reinhardtii, starting within minutes with the upregulation of alternative N assimilation routes and carbohydrate synthesis and subsequently a more gradual upregulation of genes encoding enzymes of TAG synthesis. Finally, genome resequencing analysis indicated that (1) the deletion in sta6 extends into the neighboring gene encoding respiratory burst oxidase, and (2) a commonly used STA6 strain (CC-4349) as well as the sequenced reference (CC-503) are not congenic with respect to sta6 (CC-4348), underscoring the importance of using complemented strains for more rigorous assignment of phenotype to genotype.
Figures
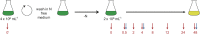
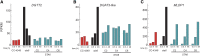

References
-
- Arrivault S., Guenther M., Ivakov A., Feil R., Vosloh D., van Dongen J.T., Sulpice R., Stitt M. (2009). Use of reverse-phase liquid chromatography, linked to tandem mass spectrometry, to profile the Calvin cycle and other metabolic intermediates in Arabidopsis rosettes at different carbon dioxide concentrations. Plant J. 59: 826–839 - PubMed
-
- Atteia A., et al. (2009). A proteomic survey of Chlamydomonas reinhardtii mitochondria sheds new light on the metabolic plasticity of the organelle and on the nature of the alpha-proteobacterial mitochondrial ancestor. Mol. Biol. Evol. 26: 1533–1548 - PubMed
-
- Ball, S., and Deschamps, P. (2009). Starch metabolism. In The Chlamydomonas Sourcebook, Vol. 2. (San Diego, CA: Academic Press).
Publication types
MeSH terms
Substances
Associated data
- Actions
- dbSNP/902873753
- dbSNP/902873754
- dbSNP/902873755
- dbSNP/902873756
- dbSNP/902873757
- dbSNP/902873758
- dbSNP/902873759
- dbSNP/902873760
- dbSNP/902873761
- dbSNP/902873762
- dbSNP/902873763
- dbSNP/902873764
- dbSNP/902873765
- dbSNP/902873766
- dbSNP/902873767
- dbSNP/902873768
- dbSNP/902873769
- dbSNP/902873770
- dbSNP/902873771
- dbSNP/902873772
- dbSNP/902873773
- dbSNP/902873774
- dbSNP/902873775
- dbSNP/902873776
- dbSNP/902873777
- dbSNP/902873778
- dbSNP/902873779
- dbSNP/902873780
- dbSNP/902873781
- dbSNP/902873782
- dbSNP/902873783
- dbSNP/902873784
- dbSNP/902873785
- dbSNP/902873786
- dbSNP/902873787
- dbSNP/902873788
- dbSNP/902873789
- dbSNP/902873790
- dbSNP/902873791
- dbSNP/902873792
- dbSNP/902873793
- dbSNP/902873794
- dbSNP/902873795
- dbSNP/902873796
- dbSNP/902873797
- dbSNP/902873798
- dbSNP/902873799
- dbSNP/902873800
- dbSNP/902873801
- dbSNP/902873802
- dbSNP/902873803
- dbSNP/902873804
- dbSNP/902873805
- dbSNP/902873806
- dbSNP/902873807
- dbSNP/902873808
- dbSNP/902873809
- dbSNP/902873810
- dbSNP/902873811
- dbSNP/902873812
- dbSNP/902873813
- dbSNP/902873814
- dbSNP/902873815
- dbSNP/902873816
- dbSNP/902873817
- dbSNP/902873818
- dbSNP/902873819
- dbSNP/902873820
- dbSNP/902873821
- dbSNP/902873822
- dbSNP/902873823
- dbSNP/902873824
- dbSNP/902873825
- dbSNP/902873826
- dbSNP/902873827
- dbSNP/902873828
- dbSNP/902873829
- dbSNP/902873830
- dbSNP/902873831
- dbSNP/902873832
- dbSNP/902873833
- dbSNP/902873834
- dbSNP/902873835
- dbSNP/902873836
- dbSNP/902873837
- dbSNP/902873838
- dbSNP/902873839
- dbSNP/902873840
- dbSNP/902873841
- dbSNP/902873842
- dbSNP/902873843
- dbSNP/902873844
- dbSNP/902873845
- dbSNP/902873846
- dbSNP/902873847
- dbSNP/902873848
- dbSNP/902873849
- dbSNP/902873850
- dbSNP/902873851
- dbSNP/902873852
- dbSNP/902873853
- dbSNP/902873854
- dbSNP/902873855
- dbSNP/902873856
- dbSNP/902873857
- dbSNP/902873858
- dbSNP/902873859
- dbSNP/902873860
- dbSNP/902873861
- dbSNP/902873862
- dbSNP/902873863
- dbSNP/902873864
- dbSNP/902873865
- dbSNP/902873866
- dbSNP/902873867
- dbSNP/902873868
- dbSNP/902873869
- dbSNP/902873870
- dbSNP/902873871
- dbSNP/902873872
- dbSNP/902873873
- dbSNP/902873874
- dbSNP/902873875
- dbSNP/902873876
- dbSNP/902873877
- dbSNP/902873878
- dbSNP/902873879
- dbSNP/902873880
- dbSNP/902873881
- dbSNP/902873882
- dbSNP/902873883
- dbSNP/902873884
- dbSNP/902873885
- dbSNP/902873886
- dbSNP/902873887
- dbSNP/902873888
- dbSNP/902873889
- dbSNP/902873890
- dbSNP/902873891
- dbSNP/902873892
- dbSNP/902873893
- dbSNP/902873894
- dbSNP/902873895
- dbSNP/902873896
- dbSNP/902873897
- dbSNP/902873898
- dbSNP/902873899
- dbSNP/902873900
- dbSNP/902873901
- dbSNP/902873902
- dbSNP/902873903
- dbSNP/902873904
- dbSNP/902873905
- dbSNP/902873906
- dbSNP/902873907
- dbSNP/902873908
- dbSNP/902873909
- dbSNP/902873910
- dbSNP/902873911
- dbSNP/902873912
- dbSNP/902873913
- dbSNP/902873914
- dbSNP/902873915
- dbSNP/902873916
- dbSNP/902873917
- dbSNP/902873918
- dbSNP/902873919
- dbSNP/902873920
- dbSNP/902873921
- dbSNP/902873922
- dbSNP/902873923
- dbSNP/902873924
- dbSNP/902873925
- dbSNP/902873926
- dbSNP/902873927
- dbSNP/902873928
- dbSNP/902873929
- dbSNP/902873930
- dbSNP/902873931
- dbSNP/902873932
- dbSNP/902873933
- dbSNP/902873934
- dbSNP/902873935
- dbSNP/902873936
- dbSNP/902873937
- dbSNP/902873938
- dbSNP/902873939
- dbSNP/902873940
- dbSNP/902873941
- dbSNP/902873942
- dbSNP/902873943
- dbSNP/902873944
- dbSNP/902873945
- dbSNP/902873946
- dbSNP/902873947
- dbSNP/902873948
- dbSNP/902873949
- dbSNP/902873950
- dbSNP/902873951
- dbSNP/902873952
- dbSNP/902873953
- dbSNP/902873954
- dbSNP/902873955
- dbSNP/902873956
- dbSNP/902873957
- dbSNP/902873958
- dbSNP/902873959
- dbSNP/902873960
- dbSNP/902873961
- dbSNP/902873962
- dbSNP/902873963
- dbSNP/902873964
- dbSNP/902873965
- dbSNP/902873966
- dbSNP/902873967
- dbSNP/902873968
- dbSNP/902873969
- dbSNP/902873970
- dbSNP/902873971
- dbSNP/902873972
- dbSNP/902873973
- dbSNP/902873974
- dbSNP/902873975
- dbSNP/902873976
- dbSNP/902873977
- dbSNP/902873978
- dbSNP/902873979
- dbSNP/902873980
- dbSNP/902873981
- dbSNP/902873982
- dbSNP/902873983
- dbSNP/902873984
- dbSNP/902873985
- dbSNP/902873986
- dbSNP/902873987
- dbSNP/902873988
- dbSNP/902873989
- dbSNP/902873990
- dbSNP/902873991
- dbSNP/902873992
- dbSNP/902873993
- dbSNP/902873994
- dbSNP/902873995
- dbSNP/902873996
- dbSNP/902873997
- dbSNP/902873998
- dbSNP/902873999
- dbSNP/902874000
- dbSNP/902874001
- dbSNP/902874002
- dbSNP/902874003
- dbSNP/902874004
- dbSNP/902874005
- dbSNP/902874006
- dbSNP/902874007
- dbSNP/902874008
- dbSNP/902874009
- dbSNP/902874010
- dbSNP/902874011
- dbSNP/902874012
- dbSNP/902874013
- dbSNP/902874014
- dbSNP/902874015
- dbSNP/902874016
- dbSNP/902874017
- dbSNP/902874018
- dbSNP/902874019
- dbSNP/902874020
- dbSNP/902874021
- dbSNP/902874022
- dbSNP/902874023
- dbSNP/902874024
- dbSNP/902874025
- dbSNP/902874026
- dbSNP/902874027
- dbSNP/902874028
- dbSNP/902874029
- dbSNP/902874030
- dbSNP/902874031
- dbSNP/902874032
- dbSNP/902874033
- dbSNP/902874034
- dbSNP/902874035
- dbSNP/902874036
- dbSNP/902874037
- dbSNP/902874038
- dbSNP/902874039
- dbSNP/902874040
- dbSNP/902874041
- dbSNP/902874042
- dbSNP/902874043
- dbSNP/902874044
- dbSNP/902874045
- dbSNP/902874046
- dbSNP/902874047
- dbSNP/902874048
- dbSNP/902874049
- dbSNP/902874050
- dbSNP/902874051
- dbSNP/902874052
- dbSNP/902874053
- dbSNP/902874054
- dbSNP/902874055
- dbSNP/902874056
- dbSNP/902874057
- dbSNP/902874058
- dbSNP/902874059
- dbSNP/902874060
- dbSNP/902874061
- dbSNP/902874062
- dbSNP/902874063
- dbSNP/902874064
- dbSNP/902874065
- dbSNP/902874066
- dbSNP/902874067
- dbSNP/902874068
- dbSNP/902874069
- dbSNP/902874070
- dbSNP/902874071
- dbSNP/902874072
- dbSNP/902874073
- dbSNP/902874074
- dbSNP/902874075
- dbSNP/902874076
- dbSNP/902874077
- dbSNP/902874078
- dbSNP/902874079
- dbSNP/902874080
- dbSNP/902874081
- dbSNP/902874082
- dbSNP/902874083
- dbSNP/902874084
- dbSNP/902874085
- dbSNP/902874086
- dbSNP/902874087
- dbSNP/902874088
- dbSNP/902874089
- dbSNP/902874090
- dbSNP/902874091
- dbSNP/902874092
- dbSNP/902874093
- dbSNP/902874094
- dbSNP/902874095
- dbSNP/902874096
- dbSNP/902874097
- dbSNP/902874098
- dbSNP/902874099
- dbSNP/902874100
- dbSNP/902874101
- dbSNP/902874102
- dbSNP/902874103
- dbSNP/902874104
- dbSNP/902874105
- dbSNP/902874106
- dbSNP/902874107
- dbSNP/902874108
- dbSNP/902874109
- dbSNP/902874110
- dbSNP/902874111
- dbSNP/902874112
- dbSNP/902874113
- dbSNP/902874114
- dbSNP/902874115
- dbSNP/902874116
- dbSNP/902874117
- dbSNP/902874118
- dbSNP/902874119
- dbSNP/902874120
- dbSNP/902874121
- dbSNP/902874122
- dbSNP/902874123
- dbSNP/902874124
- dbSNP/902874125
- dbSNP/902874126
- dbSNP/902874127
- dbSNP/902874128
- dbSNP/902874129
- dbSNP/902874130
- dbSNP/902874131
- dbSNP/902874132
- dbSNP/902874133
- dbSNP/902874134
- dbSNP/902874135
- dbSNP/902874136
- dbSNP/902874137
- dbSNP/902874138
- dbSNP/902874139
- dbSNP/902874140
- dbSNP/902874141
- dbSNP/902874142
- dbSNP/902874143
- dbSNP/902874144
- dbSNP/902874145
- dbSNP/902874146
- dbSNP/902874147
- dbSNP/902874148
- dbSNP/902874149
- dbSNP/902874150
- dbSNP/902874151
- dbSNP/902874152
- dbSNP/902874153
- dbSNP/902874154
- dbSNP/902874155
- dbSNP/902874156
- dbSNP/902874157
- dbSNP/902874158
- dbSNP/902874159
- dbSNP/902874160
- dbSNP/902874161
- dbSNP/902874162
- dbSNP/902874163
- dbSNP/902874164
- dbSNP/902874165
- dbSNP/902874166
- dbSNP/902874167
- dbSNP/902874168
- dbSNP/902874169
- dbSNP/902874170
- dbSNP/902874171
- dbSNP/902874172
- dbSNP/902874173
- dbSNP/902874174
- dbSNP/902874175
- dbSNP/902874176
- dbSNP/902874177
- dbSNP/902874178
- dbSNP/902874179
- dbSNP/902874180
- dbSNP/902874181
- dbSNP/902874182
- dbSNP/902874183
- dbSNP/902874184
- dbSNP/902874185
- dbSNP/902874186
- dbSNP/902874187
- dbSNP/902874188
- dbSNP/902874189
- dbSNP/902874190
- dbSNP/902874191
- dbSNP/902874192
- dbSNP/902874193
- dbSNP/902874194
- dbSNP/902874195
- dbSNP/902874196
- dbSNP/902874197
- dbSNP/902874198
- dbSNP/902874199
- dbSNP/902874200
- dbSNP/902874201
- dbSNP/902874202
- dbSNP/902874203
- dbSNP/902874204
- dbSNP/902874205
- dbSNP/902874206
- dbSNP/902874207
- dbSNP/902874208
- dbSNP/902874209
- dbSNP/902874210
- dbSNP/902874211
- dbSNP/902874212
- dbSNP/902874213
- dbSNP/902874214
- dbSNP/902874215
- dbSNP/902874216
- dbSNP/902874217
- dbSNP/902874218
- dbSNP/902874219
- dbSNP/902874220
- dbSNP/902874221
- dbSNP/902874222
- dbSNP/902874223
- dbSNP/902874224
- dbSNP/902874225
- dbSNP/902874226
- dbSNP/902874227
- dbSNP/902874228
- dbSNP/902874229
- dbSNP/902874230
- dbSNP/902874231
- dbSNP/902874232
- dbSNP/902874233
- dbSNP/902874234
- dbSNP/902874235
- dbSNP/902874236
- dbSNP/902874237
- dbSNP/902874238
- dbSNP/902874239
- dbSNP/902874240
- dbSNP/902874241
- dbSNP/902874242
- dbSNP/902874243
- dbSNP/902874244
- dbSNP/902874245
- dbSNP/902874246
- dbSNP/902874247
- dbSNP/902874248
- dbSNP/902874249
- dbSNP/902874250
- dbSNP/902874251
- dbSNP/902874252
- dbSNP/902874253
- dbSNP/902874254
- dbSNP/902874255
- dbSNP/902874256
- dbSNP/902874257
- dbSNP/902874258
- dbSNP/902874259
- dbSNP/902874260
- dbSNP/902874261
- dbSNP/902874262
- dbSNP/902874263
- dbSNP/902874264
- dbSNP/902874265
- dbSNP/902874266
- dbSNP/902874267
- dbSNP/902874268
- dbSNP/902874269
- dbSNP/902874270
- dbSNP/902874271
- dbSNP/902874272
- dbSNP/902874273
- dbSNP/902874274
- dbSNP/902874275
- dbSNP/902874276
- dbSNP/902874277
- dbSNP/902874278
- dbSNP/902874279
- dbSNP/902874280
- dbSNP/902874281
- dbSNP/902874282
- dbSNP/902874283
- dbSNP/902874284
- dbSNP/902874285
- dbSNP/902874286
- dbSNP/902874287
- dbSNP/902874288
- dbSNP/902874289
- dbSNP/902874290
- dbSNP/902874291
- dbSNP/902874292
- dbSNP/902874293
- dbSNP/902874294
- dbSNP/902874295
- dbSNP/902874296
- dbSNP/902874297
- dbSNP/902874298
- dbSNP/902874299
- dbSNP/902874300
- dbSNP/902874301
- dbSNP/902874302
- dbSNP/902874303
- dbSNP/902874304
- dbSNP/902874305
- dbSNP/902874306
- dbSNP/902874307
- dbSNP/902874308
- dbSNP/902874309
- dbSNP/902874310
- dbSNP/902874311
- dbSNP/902874312
- dbSNP/902874313
- dbSNP/902874314
- dbSNP/902874315
- dbSNP/902874316
- dbSNP/902874317
- dbSNP/902874318
- dbSNP/902874319
- dbSNP/902874320
- dbSNP/902874321
- dbSNP/902874322
- dbSNP/902874323
- dbSNP/902874324
- dbSNP/902874325
- dbSNP/902874326
- dbSNP/902874327
- dbSNP/902874328
- dbSNP/902874329
- dbSNP/902874330
- dbSNP/902874331
- dbSNP/902874332
- dbSNP/902874333
- dbSNP/902874334
- dbSNP/902874335
- dbSNP/902874336
- dbSNP/902874337
- dbSNP/902874338
- dbSNP/902874339
- dbSNP/902874340
- dbSNP/902874341
- dbSNP/902874342
- dbSNP/902874343
- dbSNP/902874344
- dbSNP/902874345
- dbSNP/902874346
- dbSNP/902874347
- dbSNP/902874348
- dbSNP/902874349
- dbSNP/902874350
- dbSNP/902874351
- dbSNP/902874352
- dbSNP/902874353
- dbSNP/902874354
- dbSNP/902874355
- dbSNP/902874356
- dbSNP/902874357
- dbSNP/902874358
- dbSNP/902874359
- dbSNP/902874360
- dbSNP/902874361
- dbSNP/902874362
- dbSNP/902874363
- dbSNP/902874364
- dbSNP/902874365
- dbSNP/902874366
- dbSNP/902874367
- dbSNP/902874368
- dbSNP/902874369
- dbSNP/902874370
- dbSNP/902874371
- dbSNP/902874372
- dbSNP/902874373
- dbSNP/902874374
- dbSNP/902874375
- dbSNP/902874376
- dbSNP/902874377
- dbSNP/902874378
- dbSNP/902874379
- dbSNP/902874380
- dbSNP/902874381
- dbSNP/902874382
- dbSNP/902874383
- dbSNP/902874384
- dbSNP/902874385
- dbSNP/902874386
- dbSNP/902874387
- dbSNP/902874388
- dbSNP/902874389
- dbSNP/902874390
- dbSNP/902874391
- dbSNP/902874392
- dbSNP/902874393
- dbSNP/902874394
- dbSNP/902874395
- dbSNP/902874396
- dbSNP/902874397
- dbSNP/902874398
- dbSNP/902874399
- dbSNP/902874400
- dbSNP/902874401
- dbSNP/902874402
- dbSNP/902874403
- dbSNP/902874404
- dbSNP/902874405
- dbSNP/902874406
- dbSNP/902874407
- dbSNP/902874408
- dbSNP/902874409
- dbSNP/902874410
- dbSNP/902874411
- dbSNP/902874412
- dbSNP/902874413
- dbSNP/902874414
- dbSNP/902874415
- dbSNP/902874416
- dbSNP/902874417
- dbSNP/902874418
- dbSNP/902874419
- dbSNP/902874420
- dbSNP/902874421
- dbSNP/902874422
- dbSNP/902874423
- dbSNP/902874424
- dbSNP/902874425
- dbSNP/902874426
- dbSNP/902874427
- dbSNP/902874428
- dbSNP/902874429
- dbSNP/902874430
- dbSNP/902874431
- dbSNP/902874432
- dbSNP/902874433
- dbSNP/902874434
- dbSNP/902874435
- dbSNP/902874436
- dbSNP/902874437
- dbSNP/902874438
- dbSNP/902874439
- dbSNP/902874440
- dbSNP/902874441
- dbSNP/902874442
- dbSNP/902874443
- dbSNP/902874444
- dbSNP/902874445
- dbSNP/902874446
- dbSNP/902874447
- dbSNP/902874448
- dbSNP/902874449
- dbSNP/902874450
- dbSNP/902874451
- dbSNP/902874452
- dbSNP/902874453
- dbSNP/902874454
- dbSNP/902874455
- dbSNP/902874456
- dbSNP/902874457
- dbSNP/902874458
- dbSNP/902874459
- dbSNP/902874460
- dbSNP/902874461
- dbSNP/902874462
- dbSNP/902874463
- dbSNP/902874464
- dbSNP/902874465
- dbSNP/902874466
- dbSNP/902874467
- dbSNP/902874468
- dbSNP/902874469
- dbSNP/902874470
- dbSNP/902874471
- dbSNP/902874472
- dbSNP/902874473
- dbSNP/902874474
- dbSNP/902874475
- dbSNP/902874476
- dbSNP/902874477
- dbSNP/902874478
- dbSNP/902874479
- dbSNP/902874480
- dbSNP/902874481
- dbSNP/902874482
- dbSNP/902874483
- dbSNP/902874484
- dbSNP/902874485
- dbSNP/902874486
- dbSNP/902874487
- dbSNP/902874488
- dbSNP/902874489
- dbSNP/902874490
- dbSNP/902874491
- dbSNP/902874492
- dbSNP/902874493
- dbSNP/902874494
- dbSNP/902874495
- dbSNP/902874496
- dbSNP/902874497
- dbSNP/902874498
- dbSNP/902874499
- dbSNP/902874500
- dbSNP/902874501
- dbSNP/902874502
- dbSNP/902874503
- dbSNP/902874504
- dbSNP/902874505
- dbSNP/902874506
- dbSNP/902874507
- dbSNP/902874508
- dbSNP/902874509
- dbSNP/902874510
- dbSNP/902874511
- dbSNP/902874512
- dbSNP/902874513
- dbSNP/902874514
- dbSNP/902874515
- dbSNP/902874516
- dbSNP/902874517
- dbSNP/902874518
- dbSNP/902874519
- dbSNP/902874520
- dbSNP/902874521
- dbSNP/902874522
- dbSNP/902874523
- dbSNP/902874524
- dbSNP/902874525
- dbSNP/902874526
- dbSNP/902874527
- dbSNP/902874528
- dbSNP/902874529
- dbSNP/902874530
- dbSNP/902874531
- dbSNP/902874532
- dbSNP/902874533
- dbSNP/902874534
- dbSNP/902874535
- dbSNP/902874536
- dbSNP/902874537
- dbSNP/902874538
- dbSNP/902874539
- dbSNP/902874540
- dbSNP/902874541
- dbSNP/902874542
- dbSNP/902874543
- dbSNP/902874544
- dbSNP/902874545
- dbSNP/902874546
- dbSNP/902874547
- dbSNP/902874548
- dbSNP/902874549
- dbSNP/902874550
- dbSNP/902874551
- dbSNP/902874552
- dbSNP/902874553
- dbSNP/902874554
- dbSNP/902874555
- dbSNP/902874556
- dbSNP/902874557
- dbSNP/902874558
- dbSNP/902874559
- dbSNP/902874560
- dbSNP/902874561
- dbSNP/902874562
- dbSNP/902874563
- dbSNP/902874564
- dbSNP/902874565
- dbSNP/902874566
- dbSNP/902874567
- dbSNP/902874568
- dbSNP/902874569
- dbSNP/902874570
- dbSNP/902874571
- dbSNP/902874572
- dbSNP/902874573
- dbSNP/902874574
- dbSNP/902874575
- dbSNP/902874576
- dbSNP/902874577
- dbSNP/902874578
- dbSNP/902874579
- dbSNP/902874580
- dbSNP/902874581
- dbSNP/902874582
- dbSNP/902874583
- dbSNP/902874584
- dbSNP/902874585
- dbSNP/902874586
- dbSNP/902874587
- dbSNP/902874588
- dbSNP/902874589
- dbSNP/902874590
- dbSNP/902874591
- dbSNP/902874592
- dbSNP/902874593
- dbSNP/902874594
- dbSNP/902874595
- dbSNP/902874596
- dbSNP/902874597
- dbSNP/902874598
- dbSNP/902874599
- dbSNP/902874600
- dbSNP/902874601
- dbSNP/902874602
- dbSNP/902874603
- dbSNP/902874604
- dbSNP/902874605
- dbSNP/902874606
- dbSNP/902874607
- dbSNP/902874608
- dbSNP/902874609
- dbSNP/902874610
- dbSNP/902874611
- dbSNP/902874612
- dbSNP/902874613
- dbSNP/902874614
- dbSNP/902874615
- dbSNP/902874616
- dbSNP/902874617
- dbSNP/902874618
- dbSNP/902874619
- dbSNP/902874620
- dbSNP/902874621
- dbSNP/902874622
- dbSNP/902874623
- dbSNP/902874624
- dbSNP/902874625
- dbSNP/902874626
- dbSNP/902874627
- dbSNP/902874628
- dbSNP/902874629
- dbSNP/902874630
- dbSNP/902874631
- dbSNP/902874632
- dbSNP/902874633
- dbSNP/902874634
- dbSNP/902874635
- dbSNP/902874636
- dbSNP/902874637
- dbSNP/902874638
- dbSNP/902874639
- dbSNP/902874640
- dbSNP/902874641
- dbSNP/902874642
- dbSNP/902874643
- dbSNP/902874644
- dbSNP/902874645
- dbSNP/902874646
- dbSNP/902874647
- dbSNP/902874648
- dbSNP/902874649
- dbSNP/902874650
- dbSNP/902874651
- dbSNP/902874652
- dbSNP/902874653
- dbSNP/902874654
- dbSNP/902874655
- dbSNP/902874656
- dbSNP/902874657
- dbSNP/902874658
- dbSNP/902874659
- dbSNP/902874660
- dbSNP/902874661
- dbSNP/902874662
- dbSNP/902874663
- dbSNP/902874664
- dbSNP/902874665
- dbSNP/902874666
- dbSNP/902874667
- dbSNP/902874668
- dbSNP/902874669
- dbSNP/902874670
- dbSNP/902874671
- dbSNP/902874672
- dbSNP/902874673
- dbSNP/902874674
- dbSNP/902874675
- dbSNP/902874676
- dbSNP/902874677
- dbSNP/902874678
- dbSNP/902874679
- dbSNP/902874680
- dbSNP/902874681
- dbSNP/902874682
- dbSNP/902874683
- dbSNP/902874684
- dbSNP/902874685
- dbSNP/902874686
- dbSNP/902874687
- dbSNP/902874688
- dbSNP/902874689
- dbSNP/902874690
- dbSNP/902874691
- dbSNP/902874692
- dbSNP/902874693
- dbSNP/902874694
- dbSNP/902874695
- dbSNP/902874696
- dbSNP/902874697
- dbSNP/902874698
- dbSNP/902874699
- dbSNP/902874700
- dbSNP/902874701
- dbSNP/902874702
- dbSNP/902874703
- dbSNP/902874704
- dbSNP/902874705
- dbSNP/902874706
- dbSNP/902874707
- dbSNP/902874708
- dbSNP/902874709
- dbSNP/902874710
- dbSNP/902874711
- dbSNP/902874712
- dbSNP/902874713
- dbSNP/902874714
- dbSNP/902874715
- dbSNP/902874716
- dbSNP/902874717
- dbSNP/902874718
- dbSNP/902874719
- dbSNP/902874720
- dbSNP/902874721
- dbSNP/902874722
- dbSNP/902874723
- dbSNP/902874724
- dbSNP/902874725
- dbSNP/902874726
- dbSNP/902874727
- dbSNP/902874728
- dbSNP/902874729
- dbSNP/902874730
- dbSNP/902874731
- dbSNP/902874732
- dbSNP/902874733
- dbSNP/902874734
- dbSNP/902874735
- dbSNP/902874736
- dbSNP/902874737
- dbSNP/902874738
- dbSNP/902874739
- dbSNP/902874740
- dbSNP/902874741
- dbSNP/902874742
- dbSNP/902874743
- dbSNP/902874744
- dbSNP/902874745
- dbSNP/902874746
- dbSNP/902874747
- dbSNP/902874748
- dbSNP/902874749
- dbSNP/902874750
- dbSNP/902874751
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

