The TSC-mTOR pathway regulates macrophage polarization
- PMID: 24280772
- PMCID: PMC3876736
- DOI: 10.1038/ncomms3834
The TSC-mTOR pathway regulates macrophage polarization
Abstract
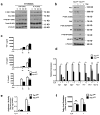
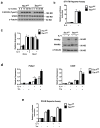
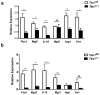
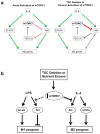
Macrophages are able to polarize to proinflammatory M1 or alternative M2 states with distinct phenotypes and physiological functions. How metabolic status regulates macrophage polarization remains not well understood, and here we examine the role of mTOR (mechanistic target of rapamycin), a central metabolic pathway that couples nutrient sensing to regulation of metabolic processes. Using a mouse model in which myeloid lineage-specific deletion of Tsc1 (Tsc1(Δ/Δ)) leads to constitutive mTOR complex 1 (mTORC1) activation, we find that Tsc1(Δ/Δ) macrophages are refractory to IL-4-induced M2 polarization, but produce increased inflammatory responses to proinflammatory stimuli. Moreover, mTORC1-mediated downregulation of Akt signalling critically contributes to defective polarization. These findings highlight a key role for the mTOR pathway in regulating macrophage polarization, and suggest how nutrient sensing and metabolic status could be 'hard-wired' to control of macrophage function, with broad implications for regulation of type 2 immunity, inflammation and allergy.
Conflict of interest statement
The authors declare no competing financial interests.
Figures


References
-
- Gordon S, Martinez FO. Alternative activation of macrophages: mechanism and functions. Immunity. 2010;32:593–604. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

