Suppression of expression between adjacent genes within heterologous modules in yeast
- PMID: 24281423
- PMCID: PMC3887525
- DOI: 10.1534/g3.113.007922
Suppression of expression between adjacent genes within heterologous modules in yeast
Abstract
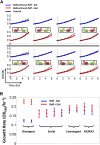
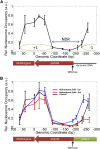
Recent studies have shown that proximal arrangement of multiple genes can have complex effects on gene expression. For example, in the case of heterologous gene expression modules, certain arrangements of the selection marker and the gene expression cassette may have unintended consequences that limit the predictability and interpretability of module behaviors. The relationship between arrangement and expression has not been systematically characterized within heterologous modules to date. In this study, we quantitatively measured gene expression patterns of the selection marker (KlURA3 driven by the promoter, pKlURA) and the gene expression cassette (GFP driven by the galactose-inducible GAL1 promoter, pGAL1) in all their possible relative arrangements in Saccharomyces cerevisiae. First, we observed that pKlURA activity depends strongly on the relative arrangement and the activity of pGAL1. Most notably, we observed transcriptional suppression in the case of divergent arrangements: pKlURA activity was reduced when pGAL1 was inactive. Based on our nucleosome occupancy data, we attribute the observed transcriptional reduction to nucleosome repositioning. Second, we observed that pGAL1 activity also depends on the relative arrangement of pKlURA. In particular, strains with divergent promoters showed significantly different pGAL1 activation patterns from other strains, but only when their growth was compromised by lack of uracil. We reasoned that this difference in pGAL1 activation patterns arises from arrangement-dependent pKlURA activity that can affect the overall cell physiology (i.e., cell growth and survival in the uracil-depleted condition). Our results underscore the necessity to consider ramifications of promoter arrangement when using synthetic gene expression modules.
Keywords: bidirectional promoter; coexpression; heterologous gene expression; nucleosome positioning.
Figures


References
-
- Acar M., Becskei A., Van Oudenaarden A., 2005. Enhancement of cellular memory by reducing stochastic transitions. Nature 435: 228–232. - PubMed
-
- Adhya S., Gottesman M., 1982. Promoter occlusion: transcription through a promoter may inhibit its activity. Cell 29: 939–944. - PubMed
-
- Bae J. Y., Laplaza J., Jeffries T. W., 2008. Effects of gene orientation and use of multiple promoters on the expression of XYL1 and XYL2 in Saccharomyces cerevisiae. Appl. Biochem. Biotechnol. 145: 69–78. - PubMed
-
- Batada N. N., Urrutia A. O., Hurst L. D., 2007. Chromatin remodelling is a major source of coexpression of linked genes in yeast. Trends Genet. 23: 480–484. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
