Quantitative exploration of the molecular origin of the activation of GTPase
- PMID: 24282301
- PMCID: PMC3870692
- DOI: 10.1073/pnas.1319854110
Quantitative exploration of the molecular origin of the activation of GTPase
Abstract
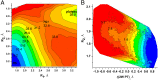
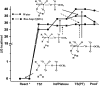
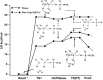
GTPases play a major role in cellular processes, and gaining quantitative understanding of their activation demands reliable free energy surfaces of the relevant mechanistic paths in solution, as well as the interpolation of this information to GTPases. Recently, we generated ab initio quantum mechanical/molecular mechanical free energy surfaces for the hydrolysis of phosphate monoesters in solution, establishing quantitatively that the barrier for the reactions with a proton transfer (PT) step from a single attacking water (1 W) is higher than the one where the PT is assisted by a second water (2 W). The implication of this finding on the activation of GTPases is quantified here, by using the ab initio solution surfaces to calibrate empirical valence bond surfaces and then exploring the origin of the activation effect. It is found that, although the 2 W PT path is a new element, this step is not rate determining, and the catalytic effect is actually due to the electrostatic stabilization of the pre-PT transition state and the subsequent plateau. Thus, the electrostatic catalytic effect found in our previous studies of the Ras GTPase activating protein (RasGAP) and the elongation factor-Tu (EF-Tu) with a 1 W mechanism is still valid for the 2 W path. Furthermore, as found before, the corresponding activation appears to involve a major allosteric effect. Overall, we believe that our finding is general to both GTPases and ATPases. In addition to the biologically relevant finding, we also provide a critical discussion of the requirements from reliable surfaces for enzymatic reactions.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
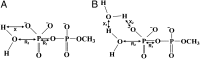


References
-
- Warshel A, Levitt M. Theoretical studies of enzymic reactions: Dielectric, electrostatic and steric stabilization of the carbonium ion in the reaction of lysozyme. J Mol Biol. 1976;103(2):227–249. - PubMed
-
- Senn HM, Thiel W. QM/MM methods for biomolecular systems. Angew Chem Int Ed Engl. 2009;48(7):1198–1229. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

