Differential effects of antibiotic therapy on the structure and function of human gut microbiota
- PMID: 24282523
- PMCID: PMC3839934
- DOI: 10.1371/journal.pone.0080201
Differential effects of antibiotic therapy on the structure and function of human gut microbiota
Abstract
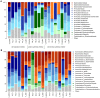
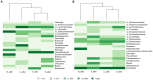
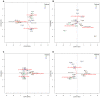
The human intestinal microbiota performs many essential functions for the host. Antimicrobial agents, such as antibiotics (AB), are also known to disturb microbial community equilibrium, thereby having an impact on human physiology. While an increasing number of studies investigate the effects of AB usage on changes in human gut microbiota biodiversity, its functional effects are still poorly understood. We performed a follow-up study to explore the effect of ABs with different modes of action on human gut microbiota composition and function. Four individuals were treated with different antibiotics and samples were taken before, during and after the AB course for all of them. Changes in the total and in the active (growing) microbiota as well as the functional changes were addressed by 16S rRNA gene and metagenomic 454-based pyrosequencing approaches. We have found that the class of antibiotic, particularly its antimicrobial effect and mode of action, played an important role in modulating the gut microbiota composition and function. Furthermore, analysis of the resistome suggested that oscillatory dynamics are not only due to antibiotic-target resistance, but also to fluctuations in the surviving bacterial community. Our results indicated that the effect of AB on the human gut microbiota relates to the interaction of several factors, principally the properties of the antimicrobial agent, and the structure, functions and resistance genes of the microbial community.
Conflict of interest statement
Figures

References
-
- Tap J, Mondot S, Levenez F, Pelletier E, Caron C, et al. (2009) Towards the human intestinal microbiota phylogenetic core. Environ Microbiol 11: 2574–2584. - PubMed
-
- Durbán A, Abellán JJ, Jiménez-Hernández N, Latorre A, Moya A (2012) Daily follow-up of bacterial communities in the human gut reveals stable composition and host-specific patterns of interaction. FEMS Microbiol Ecol 81: 427–437. - PubMed
-
- Hooper LV (2004) Bacterial contributions to mammalian gut development. Trends Microbiol 12: 129–134. - PubMed
-
- Guarner F, Malagelada JR (2003) Gut flora in health and disease. Lancet 361: 512–519. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

