A genome-wide aberrant RNA splicing in patients with acute myeloid leukemia identifies novel potential disease markers and therapeutic targets
- PMID: 24284058
- PMCID: PMC4458245
- DOI: 10.1158/1078-0432.CCR-13-0956
A genome-wide aberrant RNA splicing in patients with acute myeloid leukemia identifies novel potential disease markers and therapeutic targets
Abstract
Purpose: Despite new treatments, acute myeloid leukemia (AML) remains an incurable disease. More effective drug design requires an expanded view of the molecular complexity that underlies AML. Alternative splicing of RNA is used by normal cells to generate protein diversity. Growing evidence indicates that aberrant splicing of genes plays a key role in cancer. We investigated genome-wide splicing abnormalities in AML and based on these abnormalities, we aimed to identify novel potential biomarkers and therapeutic targets.
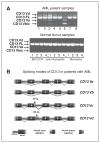
Experimental design: We used genome-wide alternative splicing screening to investigate alternative splicing abnormalities in two independent AML patient cohorts [Dana-Farber Cancer Institute (DFCI) (Boston, MA) and University Hospital de Nantes (UHN) (Nantes, France)] and normal donors. Selected splicing events were confirmed through cloning and sequencing analysis, and than validated in 193 patients with AML.
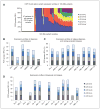
Results: Our results show that approximately 29% of expressed genes genome-wide were differentially and recurrently spliced in patients with AML compared with normal donors bone marrow CD34(+) cells. Results were reproducible in two independent AML cohorts. In both cohorts, annotation analyses indicated similar proportions of differentially spliced genes encoding several oncogenes, tumor suppressor proteins, splicing factors, and heterogeneous-nuclear-ribonucleoproteins, proteins involved in apoptosis, cell proliferation, and spliceosome assembly. Our findings are consistent with reports for other malignances and indicate that AML-specific aberrations in splicing mechanisms are a hallmark of AML pathogenesis.
Conclusions: Overall, our results suggest that aberrant splicing is a common characteristic for AML. Our findings also suggest that splice variant transcripts that are the result of splicing aberrations create novel disease markers and provide potential targets for small molecules or antibody therapeutics for this disease.
©2013 AACR
Conflict of interest statement
J.D. Griffin has commercial research grant from Jenssen Pharmaceuticals and Novartis Pharmaceuticals and is a consultant/advisory board member of Curis Pharmaceuticals and Novartis Pharmaceuticals. No potential conflicts of interest were disclosed by the other authors.
Figures

References
-
- Ast G. How did alternative splicing evolve? Nat Rev Genet. 2004;5:773–82. - PubMed
-
- Cartegni L, Chew SL, Krainer AR. Listening to silence and understanding nonsense: exonic mutations that affect splicing. Nat Rev Genet. 2002;3:285–98. - PubMed
-
- Jurica MS, Moore MJ. Pre-mRNA splicing: awash in a sea of proteins. Mol Cell. 2003;12:5–14. - PubMed
-
- Jurica MS. Searching for a wrench to throw into the splicing machine. Nat Chem Biol. 2008;4:3–6. - PubMed
-
- Hastings ML, Krainer AR. Pre-mRNA splicing in the new millennium. Curr Opin Cell Biol. 2001;13:302–9. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

