Adaptation of the length scale and amplitude of the Bicoid gradient profile to achieve robust patterning in abnormally large Drosophila melanogaster embryos
- PMID: 24284208
- PMCID: PMC3865754
- DOI: 10.1242/dev.098640
Adaptation of the length scale and amplitude of the Bicoid gradient profile to achieve robust patterning in abnormally large Drosophila melanogaster embryos
Abstract
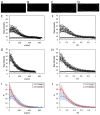
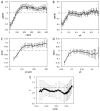
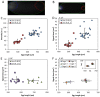
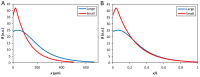
The formation of patterns that are proportional to the size of the embryo is an intriguing but poorly understood feature of development. Molecular mechanisms controlling such proportionality, or scaling, can be probed through quantitative interrogations of the properties of morphogen gradients that instruct patterning. Recent studies of the Drosophila morphogen gradient Bicoid (Bcd), which is required for anterior-posterior (AP) patterning in the early embryo, have uncovered two distinct ways of scaling. Whereas between-species scaling is achieved by adjusting the exponential shape characteristic of the Bcd gradient profile, namely, its length scale or length constant (λ), within-species scaling is achieved through adjusting the profile's amplitude, namely, the Bcd concentration at the anterior (B0). Here, we report a case in which Drosophila melanogaster embryos exhibit Bcd gradient properties uncharacteristic of their size. The embryos under investigation were from a pair of inbred lines that had been artificially selected for egg size extremes. We show that B0 in the large embryos is uncharacteristically low but λ is abnormally extended. Although the large embryos have more total bcd mRNA than their smaller counterparts, as expected, its distribution is unusually broad. We show that the large and small embryos develop gene expression patterns exhibiting boundaries that are proportional to their respective lengths. Our results suggest that the large-egg inbred line has acquired compensating properties that counteract the extreme length of the embryos to maintain Bcd gradient properties necessary for robust patterning. Our study documents, for the first time to our knowledge, a case of within-species Bcd scaling achieved through adjusting the gradient profile's exponential shape characteristic, illustrating at a molecular level how a developmental system can follow distinct operational paths towards the goal of robust and scaled patterning.
Keywords: Bicoid; Canalization; Length constant; Morphogen gradient; Robust patterning; Size scaling.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

